Abstract
Since the last century, the spread of the genotype 2 classical swine fever virus (CSFV) has caused significant issues for the pig breeding industries. Ideal strategies for controlling CSFV include vaccination and keeping farms free of CSFV. For vaccination, several attenuated CSFV viruses originating from genotype 1 are widely used; for the latter, accurate diagnosis is required for detection of the CSFV infection. Nucleic acid testing for CSFV usually uses tonsil samples, which requires an inconvenient sampling operation that injures pigs. Commercial serological tests for CSFV antibodies or antigens are unable to distinguish the genotype for originating virus. In this study, 20 mAbs were developed from the mice hybridoma cells. Four of the mAbs were identified to have the ability to only recognize the peptides derived from sub-genotype 2.1 strain, and two of them, MM1 and MM5, were further studied to identify critical binding sites (epitopes) on the E2 protein of CSFV. A total of 353 genotype 2 collections were made worldwide in GeneBank, 90.9% of which contained MM1 or MM5 epitopes. Moreover, 95.1% of sub-genotype 2.1 isolations contained MM5 epitope. Therefore, MM1 and MM5 have the potential to be developed as a diagnostic tool for detection of genotype 2 virus antigen by indirect ELISA or antibodies by competitive ELISA.
Introduction
Classical swine fever (CSF), a highly contagious swine infectious disease, hugely impacted animal health and the pig industry in the past decades (Paton and Greiser-Wilke, 2003; Blome et al., 2017). The causative agent is the classical swine fever virus (CSFV), which belongs to the Pestivirus genus within the Flaviviridae family, and is closely related to bovine viral diarrhoea virus-1 (BVDV-1), BVDV-2 and border disease virus (BDV) (Ganges et al., 2020). CSFV is an enveloped, single-stranded positive-sense RNA virus. The genome of CSFV is about 12.3 kb in length with a single open reading frame (ORF), which encodes a polyprotein that is cleaved into four structural proteins (capsid protein C, and envelope glycoproteins Erns, E1 and E2) and eight non-structural proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B). Antibodies against envelope glycoproteins E2, Erns, and NS3 can be detected in the recovering pigs from the virus infection (Fan et al., 2021). Glycoprotein E2, the most immunogenic CSFV protein, plays an important role in the induction of neutralizing antibodies and prevention of infection (Chang et al., 2010b; Velazquez-Salinas et al., 2016; Han et al., 2020).
The family of CSFV has only one serotype but three genotypes, 1, 2, and 3, and each genotype contains several sub-genotypes, sub-genotype 1.1 to 1.7, sub-genotype 2.1 to 2.7, and sub-genotype 3.1 to 3.4, respectively (Sun et al., 2013). In China, sub-genotype 2.1 has predominated since the 1990s (Gong et al., 2016; Fatima et al., 2021). The most popular vaccine in China is an attenuated C strain of CSFV or so-called “hog cholera lapinized virus” (HCLV), which belongs to CSFV genotype 1. Other routinely used vaccine such as the Lapinized Philippines Coronel (LPC) strain, or the Russian lapinized strain LK-VNIVViM also belong to CSFV genotype 1 (Chen et al., 2010; Coronado et al., 2021). Large scale vaccination of C strain over the years may result in asymptomatic transmission of other genotypes of CSFV strains in pigs (Pérez et al., 2012; Muñoz-González et al., 2015; Rios et al., 2017). While reports indicate that the C strain vaccine cannot provide complete protection against genotype 2 virus infection (Li et al., 2007; Beer et al., 2015; Zhang et al., 2015; Yoo et al., 2018; Xing et al., 2019), the vaccinated pigs showed very low viremia for genotype 2 virus, with lower pathological changes in the tissues and organs (Luo et al., 2017b), supporting the notion of asymptomatic transmission.
Establishment of CSFV-free herds or the elimination of wild type CSFV are the ideal strategy to avoid the threat of CSF in large pig farms. However, because CSFV has only one serotype, it is difficult to diagnose using normal serological methods whether the C strain-vaccinated herds have been infected with genotype 2 virus. On the other hand, direct test for virus genome with pig tonsil is usually rejected by farmers because of the inconvenient operation that injures pigs (de Smit, 2000; Wang et al., 2020a). Therefore, for timely and accurate detection and removal of the genotype 2 virus infected pigs, a differential diagnosis method distinguishing the antibodies from originating viruses needs to be developed.
In this study, we developed genotype 2 strain specific monoclonal antibodies, which can be applied for detection of either E2 antigen or antibodies from genotype 2 CSFV.
Materials and methods
Cells
The insect cell lines, Spodoptera frugiperda 9 (Sf9) and High Five were cultured at 27°C in medium IB905 (Zhejiang Yishengke Biotechnology Co., Ltd., China). Murine SP2/0-Ag14 (SP2/0) myeloma cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific Inc.) at 37°C with 10% fetal bovine serum (FBS, Thermo Fisher Scientific Inc.) and 5% CO2.
Antibodies and reagents
Horseradish peroxidase (HRP) conjugated mouse anti-His IgG or sheep anti-mouse IgG, and ×2 High-Fidelity PCR Master Mix were purchased from Sangon Biotech Co. Ltd. (Shanghai, China); SuperScript™ IV CellsDirect™ cDNA Synthesis Kit was purchased from Thermo Fisher Scientific Inc.
Preparation of the recombinant proteins
According to the instruction manual of the “Bac-to-Bac® Baculovirus Expression System” (Thermo Fisher Scientific Inc.), the genes of the recombinant proteins, E2ab-2.1 and E2ab-C (Figure 1A) were inserted respectively into the plasmid, pFastBac™1 through the restriction endonuclease sites BamHI and XhoI. These plasmids were used to transfect Sf9 cells for generation of the baculoviruses, which were used to infect High Five cells for expression of the recombinant proteins. The High Five cells were lysed and centrifuged to obtained expressed proteins, E2ab-2.1 or E2ab-C, in the pellets. The pellets were washed using a buffer solution containing 50 mM Tris, 150 mM NaCl, 3 M guanidine hydrochloride, 1 mM EDTA, and 1% Triton X-100. The washed pellets were dissolved with denaturing buffer containing 6 M guanidine hydrochloride. The proteins were recovered and concentrated by gradually removing guanidine hydrochloride with 10KD ultrafiltration tubes (Amicon Ultra-15 Centrifugal Filter, Millipore).
FIGURE 1
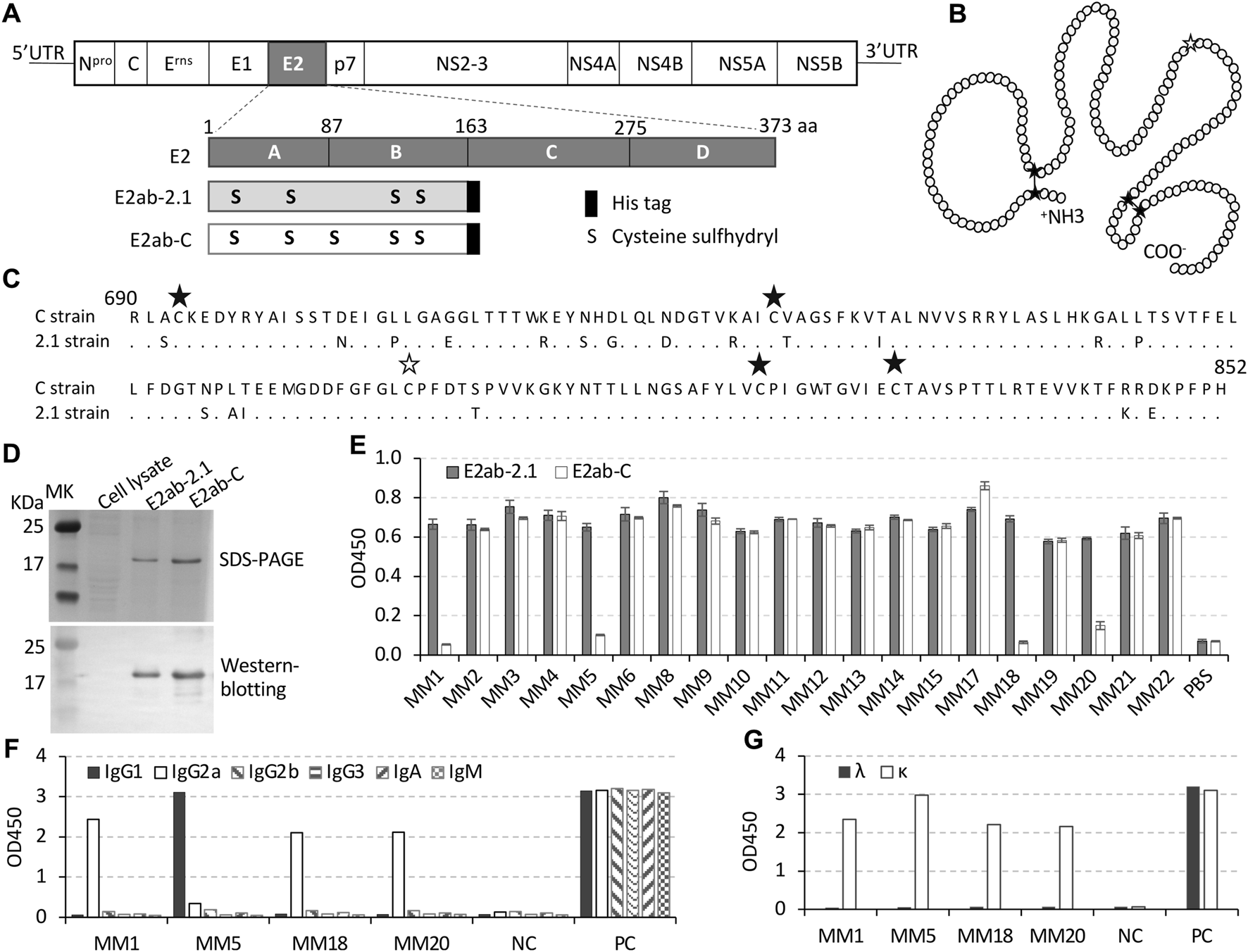
Expression of E2 recombinant protein and generation of mAbs. (A) Schematic of CSFV genome and its E2 recombinant protein. E2 protein has 373 residues, which are divided into four domains, A, B, C, and D according to the 3D structure. Recombinant protein E2ab-2.1 contains domain A and B of the E2 protein from the 2.1 strain (GeneBank accession number KU375260.1) while E2ab-C has AB domains from C strain (GeneBank accession number AF091507). The third cysteine residue in the E2 protein of the wildtype 2.1 strain was mutated to serine for generation of the recombinant E2ab-2.1 protein. Each recombinant protein was tagged with eight histidine residues at the C-terminal. (B) Cartoon diagram of disulfide bonds in natural E2 AB domains. Black asterisks indicate the cysteine residues, while white asterisk represents the cysteine mutated serine. The disulfide bonds are indicated by lines between cystine residues. (C) Alignment of two amino acid sequences of E2 AB domains from the 2.1 strain and C strain. The dot represented the conserved amino acids, the black asterisks indicate cysteine residues, and the white asterisk indicate the position of cysteine-to-serine mutation in E2ab-2.1. The numbers “690” and “853” indicate the positions of the residues in the full amino acid sequences of the CSFV polyprotein. (D) SDS-PAGE stained with Coomassie displayed purified recombinant protein E2ab-2.1 and E2ab-C, and the Western blotting indicated the proteins with His antibody. (E) Indirect ELISA was performed to detect the binding affinity of the 20 mAbs to E2ab-2.1 or E2ab-C. (F,G) The classes of heavy chains (F) and light chains (G) were detected by ELISA. Each experiment was performed in duplicate.
Generation of monoclonal antibodies
The 6 weeks-old female BALB/c mice were immunized by subcutaneous injection of the purified protein E2ab-2.1 (10 μg per mouse), which was emulsified with Freund’s complete adjuvant. After 4 weeks, the mice were injected with the same dose of the protein emulsified with Freund’s incomplete adjuvant, followed by an additional intravenous injection of the same dose after additional 3 weeks. Subsequently, another week later, hybridoma cells were produced by sacrificing the mice and fusing their spleen cells with the SP2/0 cells with the conventional method (Cho et al., 2005).
To obtain monoclonal antibodies, the hybridoma cells were cultured in 96-well plate until single cell clones were obtained. The culture supernatants were tested for the presence of antibodies against E2 using indirect ELISA. The clones expressing E2 specific antibodies were further subcloned and expanded. The mAbs were purified from the culture supernatant using protein A/G agarose beads (Beyotime Biotechnology Co., Ltd., China).
Indirect ELISA
To identify the binding sites of the mAbs on E2 protein, indirect ELISA was performed by using the different E2 proteins or peptides. Briefly, 96-well plates were coated with the protein E2ab-2.1 or E2ab-C (10 μg/mL), or chemically synthesized peptides (20 μg/mL) at 4°C overnight. The coated plates were blocked with 5% skimmed milk for 2 h at 37°C followed by incubation with the hybridoma cell culture supernatant, or purified mAbs (1 μg/mL), or pig serum (diluted 1:100) for 1 h at 37°C. The plates were washed with PBS and incubated with HRP-conjugated goat anti-mouse or anti-pig IgG secondary antibodies for 30 min at 37°C. Finally, the TMB substrate solution was added to the plates and the absorbance was measured at 450 nm. The statistic analysis of the binding affinity was performed using the OD450 values.
Identification of the classes of the mAbs
The classes of the mouse mAbs (isotypes of heavy chain and light chain) were identified using a commercial indirect ELISA kit (Wuhan Chundu Biotechnology Co., Ltd., China). The assay was performed according to the manufacturer’s instructions.
Western blotting
To check the purified recombinant proteins, the samples were separated in 10% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, United States). The membranes were blocked with 5% skimmed milk, probed with mouse anti-His antibody (1:5,000 dilution) (Sangon Biotech Co. Ltd., Shanghai, China), mAb MM1 (1 μg/mL), or mAb MM5 (1 μg/mL) followed by HRP-conjugated goat anti-mouse IgG secondary antibody (Sungene Biotechnology Co., Ltd., Shanghai, China). The enhanced chemiluminescent (ECL) reagent (NCM Biotechnology Co., Ltd., Suzhou, China) was added to capture the images with MiniChemi 610 chemiluminescent imaging system (SinSage Technology Co., Ltd., Beijing, China).
Cloning the variable regions of the mAbs
To prepare cDNA that encodes the variable regions of the mAbs, hybridoma cells were collected at the logarithmic growth phase by removing the culture medium, washing cells once with pre-cooled PBS, and resuspending with pre-cooled PBS. The cell concentration was adjusted to 2 × 106 cells/mL for mRNA isolation and cDNA synthesis according to the instruction manual of the SuperScript™ IV CellsDirect™ cDNA Synthesis Kit.
To design the amplification primers, multiple mouse immunoglobulin gene sequences were downloaded from the GenBank. The gene sequences of the variable regions were confirmed as shown in Figure 4A by aligning these sequences with each other. The GenBank codes of the sequences used for design of the heavy chain primers were X70423.1, BC092295.1, BC092271.1, BC057672.1, BC018280.1, and BC003878.1, and for light chain primers were DQ078272.1, LC522515.1, MH208237.1, LC199874.1, LC026057.1, BC080787.1, BC094013.1, BC091750.1, BC028540.1, BC019474., and BC002112.1.
The primer locations on the gene sequences were as shown in Figure 4A. Since the nucleotide sequences of the leader region within the variable regions were different between the mouse immunoglobulin gene sequences, multiple upstream primers were included along with a downstream primer in each PCR reaction. The upstream primers for the heavy chain included three oligonucleotides, Fh-1: 5′-ATGGGATGGAGCTGTATCATCC-3′, Fh-2: 5′-AGGAACTGCAGGTGTCC-3′, and Fh-3: 5′-CAGCTACAGGTGTCCACTCC-3′, with a downstream primer, Rh: 5′-AGAAGGTGTGCACACCGCTGGAC-3′. The upstream primers for the light chain included two oligonucleotides, Fκ-1: 5′-CAGTTCCTGTTTCTGTTARTGCTCTGG-3′ and Fκ-2: 5′-TGGGTGCTGCTGCTCTGGGT-3′, with a downstream primer, Rh: 5′-AGAAGGTGTGCACACCGCTGGAC-3′. The resulting PCR products were sequenced, and corresponding amino acid sequences were analyzed to confirm the framework regions (FR) and the complementary determining regions (CDR).
Competitive ELISA
Similar to the indirect ELISA, 96-well plates were coated with polypeptides 2.1-1 or 2.1-7 listed in Table 1 (7 μg/mL, 50 μL/well) at 4°C overnight. The plates were washed and incubated with 80 μL/well pig serum (1:100 dilution) followed by 100 μL/well MM5 antibody (1 μg/mL) or 100 μL/well MM12 antibody (1 μg/mL) for 1 h at 37°C. Subsequently, the plates were incubated with HRP-conjugated goat anti-mouse secondary antibody for 30 min at 37°C. The TMB substrate solution was added to the plates, and the absorbance was measured at 450 nm. The data were analyzed same as indirect ELISA.
TABLE 1
| Name | Sequence |
|---|---|
| 2.1-1 | 698RYAISSTNEIGPLGAEGLTTTW719, a |
| 2.1-2 | 716TTTWREYSHGLQLDDGTVRAICTAGSFK743 |
| 2.1-3 | 740GSFKVIALNVVSRRYLASLHKRALPTSVTFEL771 |
| 2.1-4 | 768TFELLFDGTSPAIEEMGDDFGFGLSPFDT796 |
| 2.1-5 | 779AIEEMGDDFGFGLSPFDTTPVVK801 |
| 2.1-6 | 799VVKGKYNTTLLNGSAFYLVCPIGWT823 |
| 2.1-7 | 821GWTGVIECTAVSPTTLRTEVVKTFKREKPFPH852 |
| C-1 | 700AISSTNEIGLLGAGGLTTTW719 |
| C-2 | 716TTTWKEYSQDLQLNDGTVKAICVAGSFK743 |
| C-3 | 740GSFKVTALNVVSRRYLASLHKGALLTSVTFEL771 |
| C-4 | 768TFELLFDGTNPLTEEMGDDFGFGLCPF794 |
| C-5 | 821GWTGVIECTAVSPTTLRTEVVKTFRRDKPFPH852 |
Sequences of the E2 based polypeptides used to map the binding sites for MM1 and MM5 antibodies.
The position of the residues in the amino acid sequence of the CSFV polyprotein.
Institutional review board statement
The animal studies were approved by Ethics Committee of Guangdong Haid Group Co., Ltd. (HD-211110-1, 07 December 2021).
Results
Expression of E2 recombinant proteins and generation of monoclonal antibodies
As shown in Figures 1A–C, the E2 AB domains from the 2.1 strain were selected as the immunogen. To keep a natural E2-like structure, the third cysteine was substituted with serine, which would remove possibilities of forming disulfide bonds with other cysteines in the recombinant protein (Chang et al., 2012a; Chang et al., 2012b). The immunogen was designated as E2ab-2.1, while the control protein was designated as E2ab-C, which contained E2 AB domains from the C strain without a mutation. The purified recombinant proteins E2ab-2.1 and E2ab-C were verified by Western blotting, the results indicated that these two proteins had similar molecular weight, about 20 kDa (Figure 1D).
Hybridoma cells were generated by fusion of spleen cells from E2ab-2.1 immunized mice with SP2/0 cells. Twenty hybridoma cell lines were screened after subcloning, which expressed mAbs that were named as MM1-MM6, MM8-MM15, and MM17-MM22. Indirect ELISA results showed that four mAbs, MM1, MM5, MM18, and MM20 only recognized E2ab-2.1, but not E2ab-C (Figure 1E). The class testing for these mAbs revealed that the heavy chain of MM1, MM18, and MM20 were IgG2a, while the heavy chain of MM5 was IgG1. The light chain was the κ chain for all four antibodies (Figures 1F, G). Remaining 16 mAbs recognized both the E2ab-2.1 and E2ab-C, which suggested that they were unable to distinguish whether E2-containing samples came from the 2.1 strain or the C strain of CSFV.
Mapping the binding sites of the mAbs
To determine the antigenic epitopes recognized by the mAbs, seven overlapped polypeptides for the 2.1 strain and five for the C strain were designed and synthesized (GL Biochem Ltd., Shanghai, China) based on the amino acid sequences of the respective E2 AB domains (Figure 2A). The amino acids sequences for these peptides are indicated in Table 1.
FIGURE 2
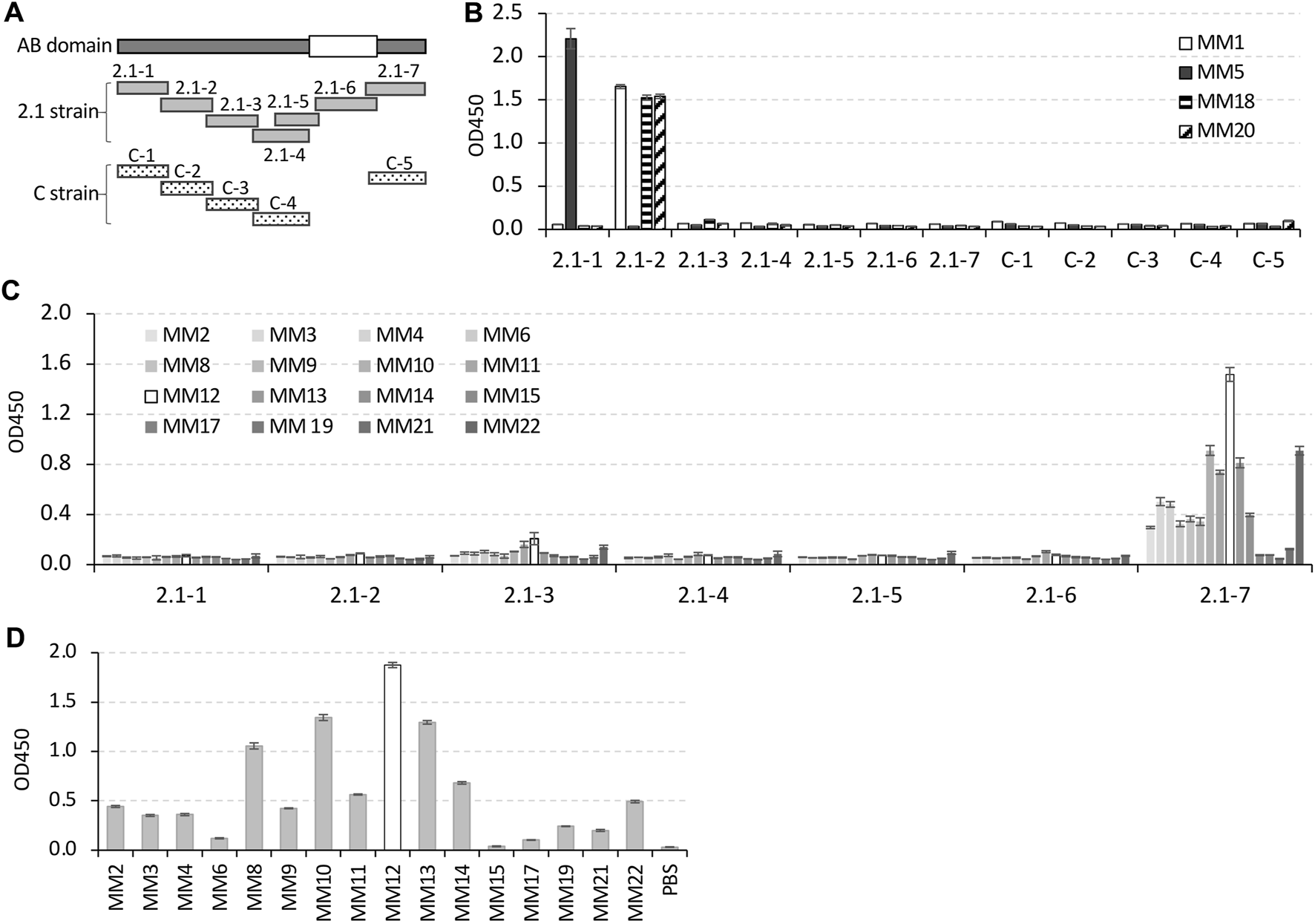
Identification of the mAbs binding sites on AB domains of the E2 protein. (A) Schematic of twelve overlapped polypeptides derived from AB domains of the E2 protein from 2.1 strain or C strain. The white square on AB domains indicates the conserved region between the 2.1 strain and C strain. The peptides derived from C strain skipped this conserved region. (B) Indirect ELISA result for binding affinity of the four 2.1 strain-specific mAbs to the twelve peptides. (C,D) Indirect ELISA results for the binding affinity of the rest 16 mAbs to 2.1 strain derived peptides (C), or to the C strain derived peptide, C-5 (D). In the histogram of (C,D), the bars for MM12 were displayed in white, and the bars for the remaining mAbs were displayed in different shades of gray (C) or gray (D).
The data from the indirect ELISA confirmed that the four 2.1 strain-specific mAbs only bound to 2.1 strain-derived polypeptides. Specifically, MM1, MM18, and MM20 only recognized peptide 2.1-2, and MM5 only recognized peptide 2.1-1 (Figure 2B). The rest of the 16 mAbs, which bound to both the 2.1 strain and C strain derived E2 recombinant proteins, recognized peptides (2.1-7 and C5) from both the strains (Figures 2C, D).
Identification of the epitopes for MM1 and MM5
Among of the 2.1 strain-specific mAbs, MM1 displayed the lowest binding affinity to C strain, and MM5 was the only mAb with the heavy chain that belonged to the IgG1 subtype (Figures 1E, F), therefore MM1 and MM5 were selected for further epitope analysis.
To determine the epitopes of MM1 and MM5, the non-conservative residues in the peptides 2.1-1 and 2.1-2 were individually mutated to the same residues as those in the C strain (Figures 3A–D). The data from ELISA demonstrated that the substitutions of the residues “709P” to “L” in peptide 2.1-1-A, or “713E” to “G” in peptide 2.1-1-B, significantly reduced the binding affinity to MM5, but no reduction in affinity was observed with the substitution of “705N” to “D” in peptide 2.1-1-D. Therefore, residues “709P” and “713E” play critical roles in the epitope of MM5, especially the residue “713E”, as the binding affinity was reduced by 95% with the E713G substitution (Figures 3A, B).
FIGURE 3
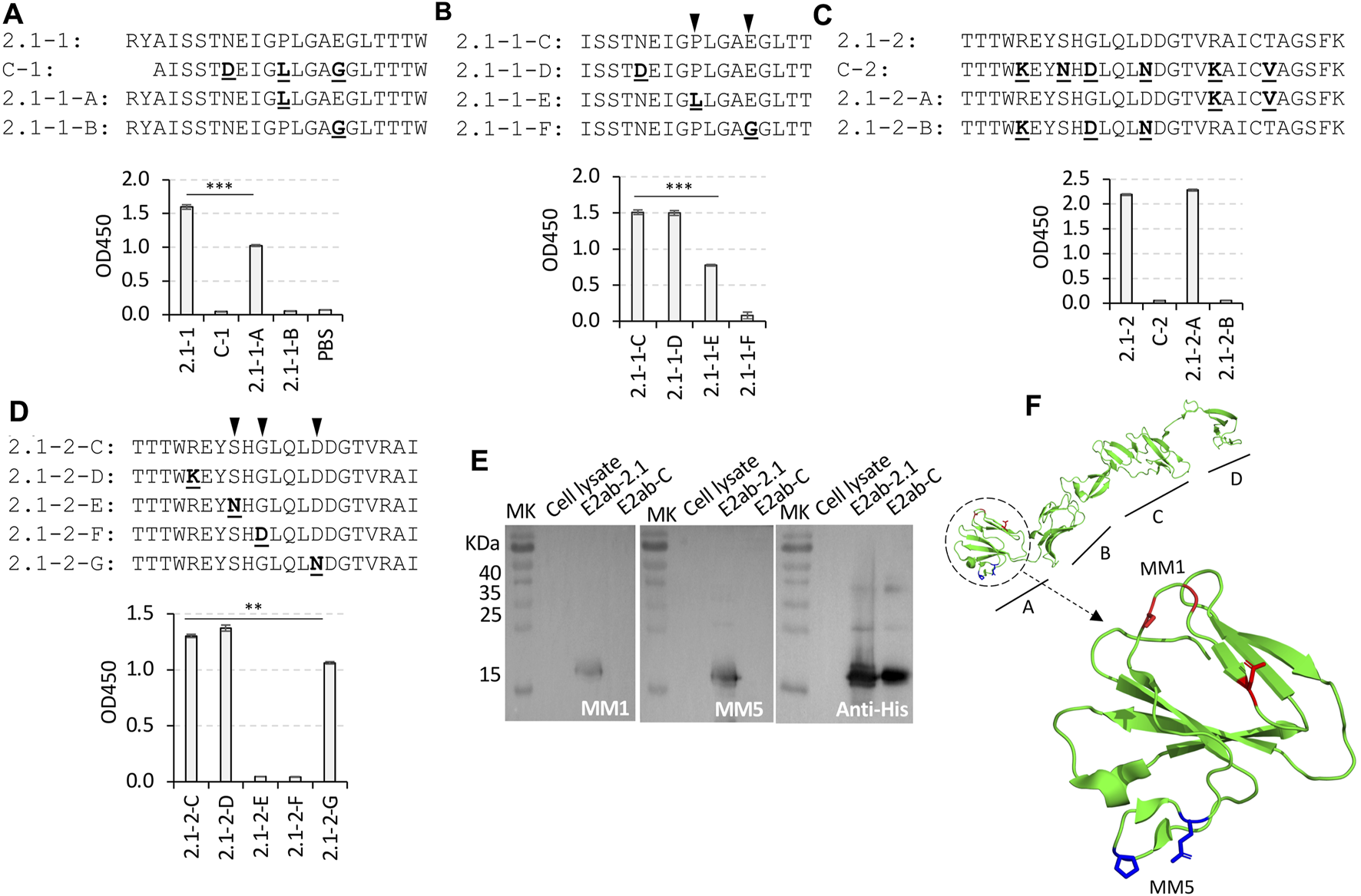
Mapping the epitope of MM1 and MM5. (A–D) A series of variant peptides synthesized based on the peptides 2.1-1 and 2.1-2 by comparison with C-1 and C-2 peptides, respectively [sequences shown in (A–D)]. Each of the non-conservative residues from the C strain peptides were used to replace the residues at the same position in the 2.1 strain peptides. The substitutions were highlighted in bold and underlined. The group of peptides originated from peptide 2.1-1 were used to map the MM5 epitope [graphs shown in (A,B)], and the group of 2.1-2 originated peptides were used to map the MM1 epitope [graphs shown in (C,D)] by ELISA. The critical binding sites of MM5 and MM1 were indicated with black inverted triangles (B,D). (E) Western blotting of protein E2ab-2.1 and E2ab-C, probed with MM1, MM5 or His antibody. (F) The 3D structure of the CSFV E2 protein displayed with the model of BVDV E2 by Pymol software (Protein Data Bank code: 2YQ2). The epitopes of MM1 and MM5 are located within the domain A, which is circled with a dotted line. In the enlarged structure of the domain A, the critical residues Serine, Glycine, and aspartate in the MM1 epitope are shown in red, while the critical residues Proline and Glutamate in the MM5 epitope are shown in blue. The experiments were performed in duplicate. **p < 0.01 compared with peptide 2.1-2-C, ***p < 0.001 compared with peptide 2.1-1 or 2.1-1-C.
Similarly, three substitution mutations (R720K, G725D, and D729N) in the variant peptide 2.1-2-B resulted in a complete loss of binding affinity to MM1 antibody. The substitutions in the last two non-conservative residues “734R” and “738T” to “K” and “V”, respectively, in the variant peptide 2.1-2-A, did not affect the binding affinity (Figure 3C).
As shown in Figure 3D, peptide 2.1-2-C was a shorter version of the peptide 2.1-2. To detect the critical residues which were responsible for MM1 recognition, four non-conservative residues in the peptide 2.1-2-C were mutated one by one to the respective residues in the C strain. The data from ELISA indicated that both the residues “723S” and “725G” were very critical in the binding of MM1 to the peptide 2.1-2-C, and replacing any of them, such as peptides 2.1-2-F or 2.1-2-F, could lead to disruption of the interaction between MM1 and peptide 2.1-2-C. Additionally, the replacement of residue “729D” with “N” (peptide 2.1-2-G) also slightly reduced the MM1 binding. These data suggest that the critical binding site for MM1 should be residues 723S, 725G and 729D on the E2 protein of the 2.1 strain.
The Western blotting results also indicated that MM1 and MM5 were able to specifically detect E2 protein derived from the 2.1 strain, but not from the C strain (Figure 3E).
The 3D model of the BVDV E2 was used to simulate structure of the CSFV E2 protein, which indicated that the locations of the MM1 and MM5 epitopes are at two different loops in the domain A (Figure 3F).
Amplification of variable region genes of the mAbs
The immunoglobulin variable region genes of the mAbs, MM1 and MM5 were amplified from the hybridoma cells (Figure 4A). Agarose gel electrophoresis results indicated that the PCR products of the heavy chains were about 550 bp and of the light chains were about 400 bp (Figure 4B). The gene fragments were sequenced and translated into amino acid sequences, from which three complementary determining regions of each the heavy chains and the light chains were identified (Figure 4C). The detail of the gene sequences and the amino acid sequences can be found in the Supplementary Data S1 (Monoclonal antibody MM1) and Supplementary Data S2 (Monoclonal antibody MM5).
FIGURE 4
Amplification of the genes that encode variable regions of mAbs, MM1 and MM5. (A) Schematic of the genes that encode variable regions of mouse immunoglobulin. The variable region genes from either heavy chain or light chain were composed of the same elements, such as, leader sequence, four FR regions, and three CDR regions. The nucleotides were numbered according to the mRNA of mouse heavy chain and light chain from Genebank (accession numbers AF466769.1 and M35669.1, respectively). The arrows indicated the primer binding positions and the gene amplification direction. (B) Gene amplifications were performed twice, and the PCR products were observed by gel electrophoresis. (C) Alignment of amino acid sequences of the heavy chains with one reference sequence (GeneBank ID CAE17334.1), and alignment of amino acid sequences of the light chains with another reference sequence (GeneBank ID CAE17333.1). The CDR area in the solid border box and the dotted border box were analyzed according to the IMGT numbering system and the Kabat numbering system, respectively (Johnson and Wu, 2000; Lefranc, 2014).
Inhibition of mAbs recognition by the specific pig serum
To further prove the recognition specificity between the mAbs and their epitopes, the pig serum was generated by immunization of pigs with the 2.1 strain derived E2 protein (Figure 5A). As shown in Figure 5B, the pig serum presented much higher binding affinities to the peptides 2.1-1 and 2.1-7 than the peptide 2.1-2. Since the peptide 2.1-1 contained the epitope of MM5, and the peptide 2.1-7 exhibited the best binding affinity to MM12 (Figures 2C, D), the binding specificity of these peptides were tested by the competitive ELISA using pig serum followed by MM5 and MM12 antibodies. The results indicated that the binding affinity of MM5 or MM12 to the peptides were significantly reduced by pre-incubation with the pig serum (Figure 5C).
FIGURE 5
The binding of the mAbs MM5 and MM12 were blocked by the pig serum. (A) The 2.1 strain specific pig serum was generated by vaccination of weaned piglets with E2ab-2.1. (B) Indirect ELISA was used to test the binding ability of the pig serum to the peptides, 2.1-1, 2.1-2, and 2.1-7. (C) The competitive ELISA was used to test the specificity of the recognition between MM5 and 2.1-1, or MM12 and 2.1-7. The immunized pig serum was used as a blocker, and the serum collected before immunization was used as a control. ***p < 0.001 compared with the serum before immunization.
Identification of MM5 and MM1 epitopes in CSFV strains
As shown in Table 2, the Supplementary Data S3 E2 sequences downloaded from GeneBank at NCBI, there were total 100 sequences for E2 protein in the genotype 1 collected from 1945 to 2021, and only one of them contained the MM5 epitope, which was isolated 30 years ago. For the genotype 2, there were in total 353 sequences for E2 protein, which belonged to seven sub-genotypes (2.1–2.7). Interestingly, 226 of 353 sequences were from sub-genotype 2.1, out of which, 215 (95.1%) sequences contained MM5 epitope. The sub-genotypes 2.2, 2.3, 2.4, 2.5, and 2.7 also had the MM5 epitope in some sequences (15.2%, 5.6%, 100%, 66.7%, and 100%, respectively), but not in sub-genotype 2.6. In addition, there were total 16 isolates in genotype 3, where only 3 of the 9 sequences from sub-genotype 3.2 had MM5 epitope.
TABLE 2
Analysis of the MM1 and MM5 epitopes in the E2 sequences.
Only the sequences which contain both MM1 and MM5 epitopes were downloaded from GenBank and analyzed. Details of the sequences were listed in the Supplementary Data S3.
Since 729D substitution has a minor effect on MM1 binding, the sequences containing the epitope of “723SHG725” were counted.
No sequence reported in China.
The genotype/subgenotype with the highest number and percentage in the each column/section were highlighted in bold.
In addition, beside the genotype 2, none of the E2 sequences from genotype 1 and genotype 3 contained the MM1 epitope. The MM1 epitope tended to exist in the sub-genotypes 2.2 and 2.3, rather than 2.1. However, if the E2 sequences from genotype 2 that contain either MM1 or MM5 epitopes counted together, the percentage of these sequences reached up to 90.8%.
The sub-genotype 2.1 predominated in world, especially in China since last 20 years (Table 2, Figure 6, Supplementary Table 1). There were total 212 sequences for E2 protein uploaded to GenBank from China from all the genotypes, where 163 (>76%) sequences belonged to sub-genotype 2.1. Out of these 163 sequences, 158 sequences (96.9%) contained the MM5 epitope. The remaining 49 sequences out of 212 were from the sub-genotypes 1.1, 2.2, 2.5, and 3.4. Notably, all the seven sequences from the sub-genotype 2.5 also contained the MM5 epitope.
FIGURE 6
Prevalence of CSFV sub-genotypes over the years worldwide (A) and in China (B) based on the E2 sequence data from the Supplementary Data S3, S4.
Moreover, it has been reported that closely related viruses, BVDV and BDV share the same epitopes with CSFV (Huang et al., 2021). However, there are only nine E2 sequences that were derived from the pig originated BVDV, and four E2 sequences from pig originated BDV in GeneBank, and the MM5 epitope was absent in the BVDV E2 sequences, but present in two of the BDV sequences. In addition, there was no MM1 epitope (SHGLQLD) present in the BDV or BVDV E2 sequences, (Supplementary Data S3 E2 sequences downloaded from GeneBank at NCBI).
Discussion
Other than directly identifying wildtype CSFV in pigs, antibodies can be identified in the serum from several days to several months or a year after infection (Clavijo et al., 2001a; Clavijo et al., 2001b; Wang et al., 2020b). There are several commercial antibody test kits available including the ones produced by IDEXX Laboratories, Inc., and Median Diagnostics, Inc., which are usually used for evaluation of the vaccine effectiveness in pigs. Most of these kits do not have abilities to distinguish the antibodies whether they are generated from the lapinized virus vaccination or from the active virus infection. As well, these kits cannot distinguish the genotype or sub-genotype of the CSFV that infected the pigs. Some test kits are used to describe special antibodies which are produced by immunizing a DIVA (Differentiating Infected from Vaccinated Animals) vaccine, such as the CSFV Erns Ab ELISA kit manufactured by Kernel Labs Inc. (Koenig et al., 2007; Gabriel et al., 2012).
Envelope protein, E2 on the surface of CSFV particle play multiple roles, such as mediation of virus infection, induction of protective immune response, or determinant of cell tropism and virulence (Risatti et al., 2005; Gavrilov et al., 2011; Liao et al., 2016; Vuono et al., 2020). The E2 protein is also a major protein in the determination of the antigenic variation of CSFV (Chen et al., 2010), and several studies on antigenic determinants with mAbs have been conducted (Chang et al., 2010a; Chang et al., 2012b; Huang et al., 2021). In this study, the AB domains of the E2 protein derived from subtype 2.1 was selected as immunogens to immunize mice, and 20 mAbs were generated. Interestingly, four mAbs, which were one-fifth of the mAbs, could distinguish 2.1 strain from the C strain. This indicated that although the E2 protein from different genotypes were highly homologous in serology, it still possessed genotype-specific antigenic determinants.
Two of 2.1 strain-specific mAbs, MM1 and MM5, were selected for further analysis. The antigenic cluster for MM1 was verified to exist in peptide 2.1-2, in which the residues serine (723S) and glycine (725G) were the critical sites for mAb binding, and both residues had small R groups. When the serine was replaced with asparagine, the interaction between MM1 and E2 was likely inhibited by the steric hindrance of the polar and larger R group of the asparagine. When the glycine was replaced with aspartate, the interaction was also inhibited likely by the negative charged R group from aspartate. In contrast, besides of residue 709P, the negatively charged glutamate (713E) present in the MM5 epitope in peptide 2.1-1, stabilizes the MM5 and E2 complex and the substitution of glutamate with glycine, a residue with a small R group, completely abolished the MM5 and E2 interaction. Furthermore, the residue 713E (strongly recognized by MM5) was reported as a critical site for the mAb T33 binding to E2 protein derived from the field strain 94.4/IL/94/TWN (subgenotype 3.4). As well, 729D (moderate recognized by MM1) was another critical site for the mAb v8 or T23 to recognize E2 protein derived from the field strain TD/96/TWN (subgenotype 2.1). Substitutions of E713G or D729N in the E2 proteins abolished the binding abilities of the mAbs T33 and T23 (Chang et al., 2010b), which indicate that the E2 A domain (Figure 2A) is the major target for generation of the subgenotype specific mAbs.
The pigs immunized with E2 protein derived from genotype 2.1 CSFV elicited fewer antibodies that recognize the MM1 epitope (Figure 5A), suggesting that the pigs would prefer generation of antibodies against MM5 epitope over the MM1 epitope. In addition, the predominant strain in recent years belonged to sub-genotype 2.1, and most of the 2.1 strains contained the MM5 epitope (Table 2), suggesting that the MM5 mAb has the potential to be developed as a differential diagnostic tool to identity 2.1 strain-specific antibody in clinical pig serum by competitive ELISA.
Moreover, genotype 2 virus has been prevailing in the world, especially in China since 2011 (Figure 6). The results from Table 2 also indicated that more than 90% of total sequences of genotype 2 isolates from worldwide contain the epitopes of MM1 and MM5 which suggests that the combination of MM1 and MM5 would likely enable detection of E2 antigens from the genotype 2 virus infected samples.
While the mAbs MM1 and MM5 have the potential to recognize the most sub-genotype 2 strains, there are still many wild strains without the epitopes for MM1 or MM5. These strains include strains from subgenotypes 2.2, 2.3, or 2.5, or from genotype 1 and 3 (Table 2). The most recent reports of the subgenotypes 2.2, 2.3, and 2.5 were in China, Latvia, and Vietnam, respectively (Postel et al., 2013a), and the last report of the subgenotype 2.2 in China was 5 years ago (Zhu et al., 2021). Due to the widely used attenuated vaccine over the years, some of the wild CSFV strains may still asymptomatically be circulating in the pig factories. In addition, international trade of live pigs and pork has the potential to introduce different subgenotypes of CSFV between countries. Therefore, it may be necessary to further develop the serologic diagnostic methods to monitor even broader subgenotypes strains.
In conclusion, MM1 and MM5 specifically recognized E2 proteins of the most prevalent CSFV genotype 2 strains. They can be developed as test reagents to detect either antigen or antibodies from genotype 2 and can be used to control CSFV infections.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by the Ethics Committee of Guangdong Haid Group Co., Ltd. (HD-211110-1, 07 December 2021). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
Conducted experiments: JZ, ZG, YZ, YY, PH, NW, ZQ, MH, JW, and LL; participated in research conception and design: CP; wrote and drafted the manuscript: JZ, ZG, ZQ, and CP; critical revision of the article: CP and JZ; performed data analysis: JZ, ZG, ZL, and CP; contributed to writing the manuscript: JZ, ZG, and CP. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Guangdong Provincial Key Laboratory of Research on the Technology of Pig-Breeding and Pig-Disease prevention (2020B1212070023).
Acknowledgments
The authors particularly thank Changchun Tu, Key Laboratory of Zoonoses Research, Ministry of Education, Zoonoses Institute, College of Veterinary Medicine, Jilin University, Changchun 130062, PR China, for his academic suggestions.
Conflict of interest
Authors JZ, ZG, YZ, YY, PH, NW, MH, CP, JW, LL, and ZL were employed by Guangdong Haid Group Co., Ltd. This work is related to a Chinese patent (ZL202111215602.5), and the author CP is the first inventor.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2023.12124/full#supplementary-material
References
1
AhujaA.BhattacharjeeU.ChakrabortyA. K.KaramA.GhatakS.PuroK.et al (2015). Complete genome sequence of classical swine fever virus subgenogroup 2.1 from Assam, India. Genome Announc.3, e01437-14. 10.1128/genomea.01437-14
2
AnD. J.LimS. I.ChoeS.KimK. S.ChaR. M.ChoI. S.et al (2018). Evolutionary dynamics of classical swine fever virus in South Korea: 1987-2017. Vet. Microbiol.225, 79–88. 10.1016/j.vetmic.2018.09.020
3
BeerM.GollerK. V.StaubachC.BlomeS. (2015). Genetic variability and distribution of classical swine fever virus. Anim. Health Res. Rev.16, 33–39. 10.1017/s1466252315000109
4
BlomeS.StaubachC.HenkeJ.CarlsonJ.BeerM. (2017). Classical swine fever-an updated review. Viruses9, 86. 10.3390/v9040086
5
ChangC. Y.HuangC. C.LinY. J.DengM. C.ChenH. C.TsaiC. H.et al (2010a). Antigenic domains analysis of classical swine fever virus E2 glycoprotein by mutagenesis and conformation-dependent monoclonal antibodies. Virus Res.149, 183–189. 10.1016/j.virusres.2010.01.016
6
ChangC. Y.HuangC. C.LinY. J.DengM. C.TsaiC. H.ChangW. M.et al (2010b). Identification of antigen-specific residues on E2 glycoprotein of classical swine fever virus. Virus Res.152, 65–72. 10.1016/j.virusres.2010.06.005
7
ChangC. Y.HuangC. C.DengM. C.HuangY. L.LinY. J.LiuH. M.et al (2012a). Antigenic mimicking with cysteine-based cyclized peptides reveals a previously unknown antigenic determinant on E2 glycoprotein of classical swine fever virus. Virus Res.163, 190–196. 10.1016/j.virusres.2011.09.019
8
ChangC. Y.HuangC. C.DengM. C.HuangY. L.LinY. J.LiuH. M.et al (2012b). Identification of conformational epitopes and antigen-specific residues at the D/A domains and the extramembrane C-terminal region of E2 glycoprotein of classical swine fever virus. Virus Res.168, 56–63. 10.1016/j.virusres.2012.06.013
9
ChenN.TongC.LiD.WanJ.YuanX.LiX.et al (2010). Antigenic analysis of classical swine fever virus E2 glycoprotein using pig antibodies identifies residues contributing to antigenic variation of the vaccine C-strain and group 2 strains circulating in China. Virol. J.7, 378. 10.1186/1743-422x-7-378
10
ChenN.HuangY.YeM.LiS.XiaoY.CuiB.et al (2019). Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018. Infect. Genet. Evol.68, 127–135. 10.1016/j.meegid.2018.12.011
11
ChoY. J.LeeD. H.KimD. O.MinW. K.BongK. T.LeeG. G.et al (2005). Production of a monoclonal antibody against ochratoxin A and its application to immunochromatographic assay. J. Agric. Food Chem.53, 8447–8451. 10.1021/jf051681q
12
ChowdryV. K.LuoY.WidénF.QiuH. J.ShanH.BelákS.et al (2014). Development of a loop-mediated isothermal amplification assay combined with a lateral flow dipstick for rapid and simple detection of classical swine fever virus in the field. J. Virol. Methods197, 14–18. 10.1016/j.jviromet.2013.11.013
13
ClavijoA.LinM.RivaJ.MalloryM.LinF.ZhouE. M. (2001a). Development of a competitive ELISA using a truncated E2 recombinant protein as antigen for detection of antibodies to classical swine fever virus. Res. Vet. Sci.70, 1–7. 10.1053/rvsc.2000.0434
14
ClavijoA.LinM.RivaJ.ZhouE. M. (2001b). Application of competitive enzyme-linked immunosorbent assay for the serologic diagnosis of classical swine fever virus infection. J. Vet. Diagn Invest.13, 357–360. 10.1177/104063870101300416
15
CoronadoL.PereraC. L.RiosL.FríasM. T.PérezL. J. (2021). A critical review about different vaccines against classical swine fever virus and their repercussions in endemic regions. Vaccines (Basel)9, 154. 10.3390/vaccines9020154
16
de SmitA. J. (2000). Laboratory diagnosis, epizootiology, and efficacy of marker vaccines in classical swine fever: a review. Vet. Q.22, 182–188. 10.1080/01652176.2000.9695054
17
FanJ.LiaoY.ZhangM.LiuC.LiZ.LiY.et al (2021). Anti-classical swine fever virus strategies. Microorganisms9, 761. 10.3390/microorganisms9040761
18
FatimaM.LuoY.ZhangL.WangP. Y.SongH.FuY.et al (2021). Genotyping and molecular characterization of classical swine fever virus isolated in China during 2016-2018. Viruses13, 664. 10.3390/v13040664
19
GabrielC.BlomeS.UrnizaA.JuanolaS.KoenenF.BeerM. (2012). Towards licensing of CP7_E2alf as marker vaccine against classical swine fever-duration of immunity. Vaccine30, 2928–2936. 10.1016/j.vaccine.2012.02.065
20
GangesL.CrookeH. R.BohórquezJ. A.PostelA.SakodaY.BecherP.et al (2020). Classical swine fever virus: the past, present and future. Virus Res.289, 198151. 10.1016/j.virusres.2020.198151
21
Garrido HaroA. D.Barrera ValleM.AcostaA.FJ. F. (2018). Phylodynamics of classical swine fever virus with emphasis on Ecuadorian strains. Transbound. Emerg. Dis.65, 782–790. 10.1111/tbed.12803
22
GavrilovB. K.RogersK.Fernandez-SainzI. J.HolinkaL. G.BorcaM. V.RisattiG. R. (2011). Effects of glycosylation on antigenicity and immunogenicity of classical swine fever virus envelope proteins. Virology420, 135–145. 10.1016/j.virol.2011.08.025
23
GongW.WuJ.LuZ.ZhangL.QinS.ChenF.et al (2016). Genetic diversity of subgenotype 2.1 isolates of classical swine fever virus. Infect. Genet. Evol.41, 218–226. 10.1016/j.meegid.2016.04.002
24
HanY.XieL.YuanM.MaY.SunH.SunY.et al (2020). Development of a marker vaccine candidate against classical swine fever based on the live attenuated vaccine C-strain. Vet. Microbiol.247, 108741. 10.1016/j.vetmic.2020.108741
25
HuangY. L.MeyerD.PostelA.TsaiK. J.LiuH. M.YangC. H.et al (2021). Identification of a common conformational epitope on the glycoprotein E2 of classical swine fever virus and border disease virus. Viruses13, 1655. 10.3390/v13081655
26
IzzatiU. Z.HoaN. T.LanN. T.DiepN. V.FukeN.HiraiT.et al (2021). Pathology of the outbreak of subgenotype 2.5 classical swine fever virus in northern Vietnam. Vet. Med. Sci.7, 164–174. 10.1002/vms3.339
27
JohnsonG.WuT. T. (2000). Kabat database and its applications: 30 years after the first variability plot. Nucleic Acids Res.28, 214–218. 10.1093/nar/28.1.214
28
KoenigP.LangeE.ReimannI.BeerM. (2007). CP7_E2alf: A safe and efficient marker vaccine strain for oral immunisation of wild boar against classical swine fever virus (CSFV). Vaccine25, 3391–3399. 10.1016/j.vaccine.2006.12.052
29
LefrancM. P. (2014). Antibody informatics: imgt, the international ImMunoGeneTics information system. Microbiol. Spectr.2. 10.1128/microbiolspec.aid-0001-2012
30
LiY.ZhaoJ. J.LiN.ShiZ.ChengD.ZhuQ. H.et al (2007). A multiplex nested RT-PCR for the detection and differentiation of wild-type viruses from C-strain vaccine of classical swine fever virus. J. Virol. Methods143, 16–22. 10.1016/j.jviromet.2007.01.032
31
LiaoX.WangZ.CaoT.TongC.GengS.GuY.et al (2016). Hypervariable antigenic region 1 of classical swine fever virus E2 protein impacts antibody neutralization. Vaccine34, 3723–3730. 10.1016/j.vaccine.2016.06.007
32
LiuC.LiM.YinX.ZhangH.XiangL.ZhaiH.et al (2018). Complete genome sequences of three sub-genotype 2.1b isolates of classical swine fever virus in China. J. Vet. Res.62, 7–15. 10.2478/jvetres-2018-0002
33
LuoY.JiS.LeiJ. L.XiangG. T.LiuY.GaoY.et al (2017a). Efficacy evaluation of the C-strain-based vaccines against the subgenotype 2.1d classical swine fever virus emerging in China. Vet. Microbiol.201, 154–161. 10.1016/j.vetmic.2017.01.012
34
LuoY.JiS.LiuY.LeiJ. L.XiaS. L.WangY.et al (2017b). Isolation and characterization of a moderately virulent classical swine fever virus emerging in China. Transbound. Emerg. Dis.64, 1848–1857. 10.1111/tbed.12581
35
MaZ.LiuM.LiuZ.MengF.WangH.CaoL.et al (2021). Epidemiological investigation of porcine circovirus type 2 and its coinfection rate in Shandong province in China from 2015 to 2018. BMC Vet. Res.17, 17. 10.1186/s12917-020-02718-4
36
Muñoz-GonzálezS.RuggliN.RosellR.PérezL. J.Frías-LeuporeauM. T.FraileL.et al (2015). Postnatal persistent infection with classical swine fever virus and its immunological implications. PLoS One10, e0125692. 10.1371/journal.pone.0125692
37
NguyenN. H.NguyenB. T. P.DoD. T.NguyenT. Q.NguyenD. T. M.NguyenM. N. (2021). Genetic diversity and molecular characterization of classical swine fever virus envelope protein genes E2 and E(rns) circulating in Vietnam from 2017 to 2019. Infect. Genet. Evol.96, 105140. 10.1016/j.meegid.2021.105140
38
PatonD. J.Greiser-WilkeI. (2003). Classical swine fever--an update. Res. Vet. Sci.75, 169–178. 10.1016/s0034-5288(03)00076-6
39
PatonD. J.McGoldrickA.Greiser-WilkeI.ParchariyanonS.SongJ. Y.LiouP. P.et al (2000). Genetic typing of classical swine fever virus. Vet. Microbiol.73, 137–157. 10.1016/s0378-1135(00)00141-3
40
PérezL. J.Díaz de ArceH.PereraC. L.RosellR.FríasM. T.PercedoM. I.et al (2012). Positive selection pressure on the B/C domains of the E2-gene of classical swine fever virus in endemic areas under C-strain vaccination. Infect. Genet. Evol.12, 1405–1412. 10.1016/j.meegid.2012.04.030
41
PostelA.SchmeiserS.BernauJ.Meindl-BoehmerA.PridotkasG.DirbakovaZ.et al (2012). Improved strategy for phylogenetic analysis of classical swine fever virus based on full-length E2 encoding sequences. Vet. Res.43, 50. 10.1186/1297-9716-43-50
42
PostelA.MoennigV.BecherP. (2013a). Classical swine fever in Europe--the current situation. Berl. Munch Tierarztl Wochenschr126, 468–475.
43
PostelA.SchmeiserS.PereraC. L.RodríguezL. J.Frias-LepoureauM. T.BecherP. (2013b). Classical swine fever virus isolates from Cuba form a new subgenotype 1.4. Vet. Microbiol.161, 334–338. 10.1016/j.vetmic.2012.07.045
44
PostelA.NishiT.KameyamaK. I.MeyerD.SuckstorffO.FukaiK.et al (2019). Reemergence of classical swine fever, Japan, 2018. Emerg. Infect. Dis.25, 1228–1231. 10.3201/eid2506.181578
45
RiosL.CoronadoL.Naranjo-FelicianoD.Martínez-PérezO.PereraC. L.Hernandez-AlvarezL.et al (2017). Deciphering the emergence, genetic diversity and evolution of classical swine fever virus. Sci. Rep.7, 17887. 10.1038/s41598-017-18196-y
46
RisattiG. R.BorcaM. V.KutishG. F.LuZ.HolinkaL. G.FrenchR. A.et al (2005). The E2 glycoprotein of classical swine fever virus is a virulence determinant in swine. J. Virol.79, 3787–3796. 10.1128/jvi.79.6.3787-3796.2005
47
SarkarS.HossainM. E.GurleyE. S.HasanR.RahmanM. Z. (2018). An outbreak of classical swine fever in pigs in Bangladesh, 2015. Vet. Med. Sci.4, 45–52. 10.1002/vms3.81
48
ShenH.PeiJ.BaiJ.ZhaoM.JuC.YiL.et al (2011). Genetic diversity and positive selection analysis of classical swine fever virus isolates in south China. Virus Genes43, 234–242. 10.1007/s11262-011-0625-5
49
SilvaM. N.SilvaD. M.LeiteA. S.GomesA. L.FreitasA. C.Pinheiro-JuniorJ. W.et al (2017). Identification and genetic characterization of classical swine fever virus isolates in Brazil: a new subgenotype. Arch. Virol.162, 817–822. 10.1007/s00705-016-3145-8
50
SunS. Q.YinS. H.GuoH. C.JinY.ShangY. J.LiuX. T. (2013). Genetic typing of classical swine fever virus isolates from China. Transbound. Emerg. Dis.60, 370–375. 10.1111/j.1865-1682.2012.01346.x
51
Velazquez-SalinasL.RisattiG. R.HolinkaL. G.O'DonnellV.CarlsonJ.AlfanoM.et al (2016). Recoding structural glycoprotein E2 in classical swine fever virus (CSFV) produces complete virus attenuation in swine and protects infected animals against disease. Virology494, 178–189. 10.1016/j.virol.2016.04.007
52
VuonoE. A.Ramirez-MedinaE.AzzinaroP.BerggrenK. A.RaiA.PruittS.et al (2020). SERTA domain containing protein 1 (SERTAD1) interacts with classical swine fever virus structural glycoprotein E2, which is involved in virus virulence in swine. Viruses12, 421. 10.3390/v12040421
53
WangL.MaderaR.LiY.McVeyD. S.DroletB. S.ShiJ. (2020a). Recent advances in the diagnosis of classical swine fever and future perspectives. Pathogens9, 658. 10.3390/pathogens9080658
54
WangL.MiS.MaderaR.GangesL.BorcaM. V.RenJ.et al (2020b). A neutralizing monoclonal antibody-based competitive ELISA for classical swine fever C-strain post-vaccination monitoring. BMC Vet. Res.16, 14. 10.1186/s12917-020-2237-6
55
XingC.LuZ.JiangJ.HuangL.XuJ.HeD.et al (2019). Sub-subgenotype 2.1c isolates of classical swine fever virus are dominant in Guangdong province of China, 2018. Infect. Genet. Evol.68, 212–217. 10.1016/j.meegid.2018.12.029
56
YooS. J.KwonT.KangK.KimH.KangS. C.RichtJ. A.et al (2018). Genetic evolution of classical swine fever virus under immune environments conditioned by genotype 1-based modified live virus vaccine. Transbound. Emerg. Dis.65, 735–745. 10.1111/tbed.12798
57
ZhangH.LengC.FengL.ZhaiH.ChenJ.LiuC.et al (2015). A new subgenotype 2.1d isolates of classical swine fever virus in China, 2014. Infect. Genet. Evol.34, 94–105. 10.1016/j.meegid.2015.05.031
58
ZhuX.LiuM.WuX.MaW.ZhaoX. (2021). Phylogenetic analysis of classical swine fever virus isolates from China. Arch. Virol.166, 2255–2261. 10.1007/s00705-021-05084-0
Summary
Keywords
classical swine fever virus, monoclonal antibodies, genotype 2, differential diagnosis, epitopes
Citation
Zhang J, Guo Z, Zhao Y, Yang Y, Huang P, Wang N, Qian Z, He M, Wu J, Luo L, Li Z and Pan C (2023) Identification of novel monoclonal antibodies specific for the conserved epitopes in the E2 protein of genotype 2 classical swine fever virus: implication for differential diagnosis. Acta Virol. 67:12124. doi: 10.3389/av.2023.12124
Received
07 February 2023
Accepted
11 September 2023
Published
11 October 2023
Volume
67 - 2023
Edited by
Katarina Polcicova, Slovak Academy of Sciences, Slovakia
Updates
Copyright
© 2023 Zhang, Guo, Zhao, Yang, Huang, Wang, Qian, He, Wu, Luo, Li and Pan.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chungen Pan, pancg01@haid.com.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.