Abstract
Despite the urgent need for effective antivirals against SARS-CoV-2 to mitigate the catastrophic impact of the COVID-19 pandemic, favipiravir and ivermectin are among the common repurposed drugs that have been provisionally used in some countries. There have been clinical trials with mixed results, and therefore, it is still inconclusive whether they are effective or should be dismissed. It is plausible that the lack of clear-cut clinical benefits was due to the finding of only marginal levels of in vivo antiviral activity. An obvious way to improve the activity of antivirals is to use them in synergistic combinations. The in vitro antiviral activity of the combinations of favipiravir, ivermectin, niclosamide, and chloroquine against SARS-CoV-2 was assessed in Vero E6 cells and the lung epithelial cell, Calu-3. Here we show that favipiravir and ivermectin had synergistic effects against SARS-CoV-2 in Vero E6 cells. In addition, we found that favipiravir had an additive effect with niclosamide, another repurposed anti-parasitic drug with anti-SARS-CoV-2 activity. However, the anti-SARS-CoV-2 activity of favipiravir was drastically reduced when evaluated in Calu-3 cells. This suggested that this cell type might not be able to metabolize favipiravir into its active form and that this deficiency in some cell types may affect the in vivo efficacy of this drug. Favipiravir and ivermectin show the best synergistic effect. This combination is being tested in a randomized controlled clinical trial (NCT05155527).
Introduction
Up to the end of June 2022, the COVID-19 pandemic had resulted in more than 6 million deaths (Worldometer, 2022). The emergence of variants with antigenic changes causing vaccine escape has obliterated the hope to eradicate the virus, and the disease will likely continue to be a major problem for the foreseeable future. Effective antivirals are urgently needed to mitigate the disease burden, especially where vaccine supplies are insufficient. In addition, infection and illnesses continue even in a vaccinated population, and effective antivirals would ensure the return to normalcy with a further reduced risk of severe disease and death. While new effective antivirals have been developed (Wen et al., 2022), they are costly and not widely available. Furthermore, treatment using monoclonal antibodies can be effective only in the early phase of the disease (Hurt and Wheatley, 2021). Repurposed drugs with antiviral activity have been the main target for developing COVID-19 treatments (Alam et al., 2021). Remdesivir is the repurposed antiviral with the most supporting clinical data for its effectiveness and is recommended by many treatment guidelines (Ansems et al., 2021; Wang et al., 2021a). However, the drug cannot be taken orally and, therefore, is not used in mild cases or in an early phase. Early antiviral treatment is very likely to be more effective and will prevent not only death but also progression to severe disease (Ngo et al., 2021). Several repurposed drugs with in vitro antiviral activity have been tested in clinical trials (Alam et al., 2021). Some have been shown to be ineffective, and some have shown mixed results. Marginal or low levels of efficacy could be a reason for the lack of a clear-cut benefit for some of these drugs.
Favipiravir has been approved in Japan as an alternative drug for the treatment of resistant influenza infections (Delang et al., 2018; Goldhill et al., 2021). Favipiravir (T-705; 6-fluoro-3-hydroxy-2-pyrazinecarboxamide) is a pyrazine analog that can prevent RNA elongation by viral RNA-dependent RNA polymerase (RdRp) (Furuta et al., 2017). The catalytic domain of RdRp is conserved among several RNA viruses; thus, favipiravir shows inhibitory effects against a broad range of RNA viruses, including influenza A viruses (Baranovich et al., 2013), respiratory syncytial virus (Jochmans et al., 2016), West Nile virus (Escribano-Romero et al., 2017), Zika virus (Kim et al., 2018; Pires de Mello et al., 2018), and Ebola virus (Guedj et al., 2018). Favipiravir is metabolized intracellularly to its active form, favipiravir-ribofuranosyl-5′-triphosphate (favipiravir-RTP) (Furuta et al., 2017). It is believed to inhibit viral replication by inhibiting viral RdRp and by inducing lethal hypermutations (Baranovich et al., 2013; Wang et al., 2021b). Recently, it was shown that favipiravir also inhibits SARS-CoV-2 by inducing lethal hypermutations (Shannon et al., 2020). The broad-spectrum antiviral activity of favipiravir makes it a potentially promising drug for the treatment of the recently emerged SARS-CoV-2. However, favipiravir was tested in clinical trials with mixed results (Hassanipour et al., 2021). It is being used for COVID-19 treatment in some countries, including Thailand (Department of Medical Services, 2020).
Ivermectin is a derivative of avermectin (a macrocyclic lactone found in Streptomyces avermectinius) and is being used for anti-parasite medication by blocking the transmission of neuronal signals of the parasites. Ivermectin was previously reported to have broad-spectrum antiviral activity against various types of RNA viruses. The proposed mechanism of action of ivermectin is that the drug molecule targets the host nuclear transport proteins, the importin α/β1 heterodimer, thereby inhibiting the nuclear import of various viral proteins. Several studies demonstrated the antiviral activity of ivermectin in flaviviruses, including dengue virus, West Nile virus, and Zika virus, by inhibiting the nuclear import of the NS5 protein (Xu et al., 2018; Kongmanas et al., 2020; Yang et al., 2020). In human immunodeficiency virus type 1 (HIV-1) and influenza A viruses, it was shown that ivermectin inhibited integrase and viral ribonucleoprotein nuclear import, respectively (Wagstaff et al., 2012; Götz et al., 2016). However, the exact mechanism of the antiviral activity of ivermectin has not yet been well described. Ivermectin also showed anti-SARS-CoV-2 activity (Caly et al., 2020; Yang et al., 2020). Although many clinical trials have shown its efficacy in COVID-19 treatment and prophylaxis (Ahmed et al., 2021; Behera et al., 2021; Kory et al., 2021; Lima-Morales et al., 2021), many others have indicated that it has no clinical benefit (Popp et al., 2021; Vallejos et al., 2021; Lim et al., 2022). These trials varied in the dosages, time of initiating the treatment, and whether to take the drug with a meal. As earlier treatment is more likely to be effective, taking ivermectin with a fatty meal was shown to increase absorption and drug plasma levels (Guzzo et al., 2002). It is still possible that these factors might negatively influence trial outcomes, and the drug may provide some benefit in specific settings despite the disappointing clinical trial results.
Niclosamide is an anthelmintic drug widely used in humans to treat tapeworm infections. The proposed mechanism involves inhibiting oxidative phosphorylation and stimulating adenosine triphosphatase activity in the mitochondria of the tapeworm (Kadri et al., 2018). From several repurposed drug screenings, niclosamide was identified as a multifunctional drug due to its ability to regulate multiple pathways, including mTORC1, STAT3, NF-κB, Notch, NS2B-NS3 interaction, and pH (Li et al., 2014; Xu et al., 2020). This made it a potential candidate for the treatment of cancer, bacterial, and viral infections. Many studies demonstrated the antiviral activity of niclosamide in several viruses, such as the Middle East respiratory syndrome coronavirus (MERS-CoV) (Gassen et al., 2019), dengue virus (Kao et al., 2018), Japanese encephalitis virus (Fang et al., 2013), hepatitis C virus (Mazzon et al., 2019), Epstein–Barr virus (Huang et al., 2017), HIV-1 (Niyomdecha et al., 2020), and the recently emerged SARS-CoV-2 (Jeon et al., 2020; Backer et al., 2021). Niclosamide is still an interesting drug that is being tested in clinical trials (Backer et al., 2021).
Chloroquine is an aminoquinoline and is primarily used for malaria treatment. Repurposed drug screening revealed antiviral activity of chloroquine in diverse types of viruses, including flaviviruses, retroviruses, and coronaviruses, by inhibiting the pH-dependent steps of viral replication (Kamat and Kumari, 2021). Many studies have shown the potent in vitro activity of chloroquine against SARS-CoV-2 (Vincent et al., 2005; Musa, 2020; Wang et al., 2020). However, chloroquine has been dismissed as ineffective in COVID-19 treatment (Singh et al., 2021).
The use of these repurposed drugs in COVID-19 was not supported by data from large clinical trials, and they are not recommended by the World Health Organization (WHO) or the US Food and Drug Administration (US FDA) (Axfors et al., 2021; US FDA, 2021; WHO, 2022). The main reason for the lack of clinical efficacy despite the in vitro activity is that the in vitro IC50 values are usually too high to be achievable in vivo at a normal therapeutic dose (Schmith et al., 2020). Increasing the dosage is not a viable solution, as it may result in toxic effects and adverse events. An approach to decreasing the IC50 values and making them lower than plasma concentrations at a normal therapeutic dose is drug combination (Phougat et al., 2014). However, synergy, or at least an additive effect, is needed to make a combination more effective than single drugs and markedly reduce the IC50s. We, therefore, tested combinations of common repurposed drugs with anti-SARS-CoV-2 activity to identify combinations with good potential for further clinical testing. These drugs are common, inexpensive, and widely available, and their combinations may provide another affordable solution for COVID-19 treatment.
Methods
Chemicals
The compounds niclosamide (N3510, Sigma), ivermectin (I8898, Sigma), and favipiravir (HY-14768, MedChemExpress) were dissolved in 100% dimethyl sulfoxide (DMSO, Sigma), and chloroquine (HY-17589, MedChemExpress) was dissolved in water and stored at −80°C. All drugs were diluted to the working concentrations in a Minimum Essential Medium with 1.5 g/L sodium bicarbonate, non-essential amino acids, L-glutamine, and sodium pyruvate (MEM; 10-009-CV, Corning) and supplemented with 2% heat-inactivated fetal bovine serum (FBS, Gibco) for the antiviral experiment in Vero E6 cells or Dulbecco’s Modified Eagle Medium/Nutrient Mixture F12 (DMEM/F12; 11320033, Gibco) supplemented with 2% heat-inactivated FBS for Calu-3 cells. The final concentration of DMSO in all experiments was 0.5%.
Cells and viruses
Vero E6 (Vero C1008, Cat. no. CRL-1586, ATCC, USA) cells were cultivated in MEM supplemented with 10% heat-inactivated FBS (10% FBS-MEM) at 37°C with 5% CO2. Calu-3 cells (Cat. no. HTB-55, ATCC, USA) were cultivated in DMEM/F12 supplemented with 10% heat-inactivated FBS (10% FBS-DMEM/F12) at 37°C with 5% CO2.
SARS-CoV-2/01/human/Jan2020/Thailand, which represented the original Wuhan strain, was isolated from nasopharyngeal swabs of a confirmed COVID-19 case in Thailand in the previous study (GenBank accession no. QYZ85362.1) (Kanjanasirirat et al., 2020). The virus was propagated in Vero E6 cells. Briefly, Vero E6 were seeded in T75 cell culture flasks at a density of 4 × 106 cells per flask before the day of infection. Then the culture medium was removed, and the cells were inoculated with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01 for 1 h. Subsequently, the viruses were discarded, and the cells were further maintained in 2% FBS-MEM at 37°C with 5% CO2. The cytopathic effects (CPE) were observed daily until the CPE reached 50%. The supernatants containing viruses were harvested by centrifugation to remove cell debris and stored at −80°C. The viral titer was determined by plaque assay in Vero E6 cells or a 50% tissue culture infectious dose (TCID50) endpoint dilution assay in Calu-3 cells.
Cell viability assay
Vero E6 or Calu-3 cells were seeded in 96-well plates at a density that allowed 100% and 70% confluence, respectively. The culture medium was removed, and the cells were treated with various concentrations of drugs for 48 h. The cell viability was assessed using MTT dyes (Invitrogen). The viable cells would convert the 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide to MTT formazan. Then 50 µL DMSO was added to dissolve the precipitates of MTT formazan in the cells, and the absorbance was read at 570 nm. The cells treated with 0.5% DMSO were used as a cell control. The 50% cytotoxic concentration (CC50) was calculated using non-linear regression analysis. The experiments were repeated at least twice (Supplementary File S2).
Drug treatments
Evaluation of single-drug treatment against SARS-CoV-2 in vitro
Vero E6 or Calu-3 cells were seeded in 96-well plates at a density of 2.5 × 104 cells per well or 2.0 × 104 cells per well, respectively, before the day of infection. Each drug was twofold serially diluted in 2% FBS-MEM or 2% FBS-DMEM/F12. The initial concentrations of favipiravir, ivermectin, niclosamide, and chloroquine used started at 1000, 40, 0.5, and 160 μM, respectively, for the treatments in Vero E6 cells. For Calu-3 cells, the initial concentrations of favipiravir and ivermectin used started at 2000 and 12.5 µM, respectively. Then the cells were pretreated with the serially diluted drugs or 0.5% DMSO as a no-drug control for 1 h at 37°C with 5% CO2. Subsequently, the cells were incubated with SARS-CoV-2 at MOI 0.01 in Vero E6 cells or 500 TCID50/100 µL in Calu-3 cells for 1 h. The optimization of SARS-CoV-2 infection in Calu-3 cells is shown in S1 Fig. After that, the viral inoculum was removed. The cells were further maintained in the media containing serially diluted drugs or 0.5% DMSO for 2 days. The virus supernatants were collected for titration using a plaque assay and one-step quantitative reverse-transcription PCR (qRT-PCR). The half-maximal inhibitory concentration (IC50) was calculated from the dose-response curve of drug treatment against SARS-CoV-2 using non-linear regression analysis. The experiments were repeated at least three times (Supplementary File S2).
Evaluation of two-drug combinations for treatment against SARS-CoV-2 in vitro
Vero E6 or Calu-3 cells were seeded in 96-well plates at a density of 2.5 × 104 cells per well or 2.0 × 104 cells per well, respectively, before the day of infection. The cells were pretreated for 1 h with 16 different pairwise combinations of two drugs. Drug concentrations ranged between 2×, 1×, 0.5× and 0.25× of the IC50 values that were determined from a single drug treatment. In Vero E6 cells, the concentration of favipiravir was 80, 40, 20, and 10 μM; ivermectin was 2.4, 1.2, 0.6, and 0.3 µM; niclosamide was 0.09, 0.045, 0.0225, and 0.01125 µM; and chloroquine was 1.7, 0.85, 0.425, and 0.2125 µM. In Calu-3 cells, the concentration of favipiravir was 1.8, 0.9, 0.45, and 0.225 mM, and the concentration of ivermectin was 0.4, 0.2, 0.1, and 0.05 µM. Subsequently, the cells were infected with SARS-CoV-2 following the same approach used in the single drug experiments. The virus supernatants were collected for titration by one-step qRT-PCR and plaque assay. The experiments were repeated at least three times (Supplementary File S2).
Viral quantitation
Plaque assay
Vero E6 cells were seeded in 12-well plates at a density of 2.2 × 105 cells per well, which allowed 100% confluence to be reached within 18 h. The culture medium was removed, and the cells were inoculated with 100 µL of 10-fold serial dilutions of virus supernatants for 1 h at 37°C with 5% CO2. After that, the virus supernatants were removed, and the cells were overlaid with 1.56% microcrystalline cellulose (Avicel, RC-591) in 2% FBS-MEM. The cells were further incubated at 37°C with 5% CO2 for 3 days. The overlaid medium was removed, and the cells were fixed with 10% (v/v) formalin in phosphate-buffered saline (PBS) for 2 h. The fixed infected cells were washed in tap water, stained with 1% (w/v) crystal violet in 20% (v/v) ethanol for 5 min, and washed to remove the excess dyes. The plaques were counted, and the viral titers were calculated in plaque-forming units per ml (pfu/mL).
50% cell culture infectious dose (TCID50) endpoint dilution assay
Calu-3 cells were seeded in 96-well plates at a density of 2 × 104 cells per well. The culture medium was removed, and the cells were incubated with a half-log10 serial dilution of the virus stock for 48 h at 37°C with 5% CO2. After that, the cells were fixed with 1:1 methanol/acetone for 30 min at 4°C and the infectivity was detected with an antibody against the SARS-CoV-2 nucleocapsid protein (40143-R001, Sino Biological). The viral TCID50 titers were calculated using the Reed and Muench method (Reed and Muench, 1938).
One-step qRT-PCR for SARS-CoV-2
The one-step qRT-PCR was used as a screening method to detect the RNA of SARS-CoV-2 directly from the virus supernatants without RNA extraction (Ganguly et al., 2020; Jitobaom et al., 2022). This assay was validated by comparing the results with viral quantitation using the plaque assay. The virus supernatants were heat-inactivated at 70°C for 20 min. Then the inactivated samples were diluted 1:10 with DNase/RNase free distilled water. One-step qRT-PCR was performed using the Power SYBR one-step kit (Applied Biosystems) and the LightCycler 480 (Roche, LC480). The one-step RT-PCR master mix was prepared following the kit’s instructions for a 10 µL reaction volume.
The primers used were N-Fw: 5′-GGGGAACTTCTCCTGCTAGAAT-3′ and N-Rv: 5′-CAGACATTTTGCTCTCAAGCTG-3′. Then 5.4 µL of the master mix was mixed with 4.6 µL of the sample in 96-well white PCR plates. RNA of SARS-CoV-2 purified from the virus stock using TRIzol-LS (Invitrogen) was used as a positive control. The samples also included a no-template control (nuclease-free water and the medium of SARS-CoV-2 mock-infected cells). The LC480 was run according to the Power SYBR one-step kit’s instructions. Briefly, the revere transcription step at 48°C for 30 min and the activation of polymerase at 95°C for 10 min was followed by 45 amplification cycles (95°C for 15s, 60°C for 1 min) and a melting curve step to determine the specificity of the PCR product from the melting temperature (Tm) (95°C for 30s, 60°C for 30s). The threshold cycle (Ct) values were calculated from raw fluorescence data using the Abs Quant/2nd derivative method. The Tm calling analysis was performed to exclude reactions with non-specific amplification by comparing no template control with the product amplified from the positive control. The inhibition of SARS-CoV-2 production in drug-treated cells was relative to the cells treated with 0.5% DMSO.
Evaluation of the combination synergy scores
The SynergyFinderPlus web application was used to analyze and visualize the degree of combination synergy (Zheng et al., 2022). The synergy scores of two-drug combinations were analyzed by comparing the observed drug combination response (percent inhibition) against the expected response, calculated using reference models (Ianevski et al., 2020). The reference models used in this study include the Loewe additivity (Loewe), Zero Independence Potency (ZIP), Highest Single Agent (HSA), and Bliss independence models (Bliss) (Yadav et al., 2015). For a synergy score less than −10, the interaction between two drugs accounted for antagonistic effects; from −10 to 10, the interaction between two drugs accounted for additive effects; and larger than 10, the interaction between two drugs accounted for synergistic effects.
Statistical analysis
All drug treatment experiments were performed in triplicate, and the data are shown as mean ± SD. The IC50 and the CC50 were calculated by non-linear regression analysis using GraphPad Prism 9 (GraphPad Software, Inc., CA). The selectivity index (SI) was calculated from the ratio between CC50 and IC50.
Results
Single drug treatment against SARS-CoV-2 in Vero E6 cells
The main features of the antiviral activities of favipiravir and the repurposed anti-parasitic drugs niclosamide, ivermectin, and chloroquine are shown in Table 1 and Figure 1. Virus production was determined using both plaque assays and one-step qRT-PCR and is expressed as the percent inhibition relative to the DMSO-treated cell control. The IC50 values determined by both methods are similar. The IC50 values calculated from the dose-response determined by one-step qRT-PCR for favipiravir, ivermectin, niclosamide, and chloroquine were 40.49, 1.24, 0.048, and 0.89 µM, respectively. And the IC50 values calculated from the dose-response determined by plaque assay for favipiravir, ivermectin, niclosamide, and chloroquine were 41.81, 1.23, 0.046, and 0.82 µM, respectively. This confirmed that viral RNA quantitation could accurately determine the infectious viral output for these experiments, and for higher throughput, one-step qRT-PCR was used for screening of two-drug combinations. The selectivity indexes of each single drug treatment are also shown in Table 1.
TABLE 1
| Drug candidates | Drug class | Drug indication | CC50 (µM) | IC50 (µM) qRT-PCR | IC50 (µM) plaque assay | SI (CC50/IC50) |
|---|---|---|---|---|---|---|
| Favipiravir | Direct antiviral agents | Treatment of influenza, and other viral infections | 501.40 | 40.49 | 41.81 | 12.18 |
| Ivermectin | Anti-parasitic agents | Treatment of onchocerciasis, and other worm infestations | 10.55 | 1.24 | 1.23 | 8.54 |
| Niclosamide | Anti-parasitic agents | Treatment of tapeworm and intestinal fluke infections | 0.29 | 0.048 | 0.046 | 6.17 |
| Chloroquine | Anti-malarial agents | Treatment of malaria, rheumatic diseases and Zika virus infection | 122.10 | 0.89 | 0.82 | 142.81 |
The single drug treatment against SARS-CoV-2 in Vero E6 cells.
FIGURE 1
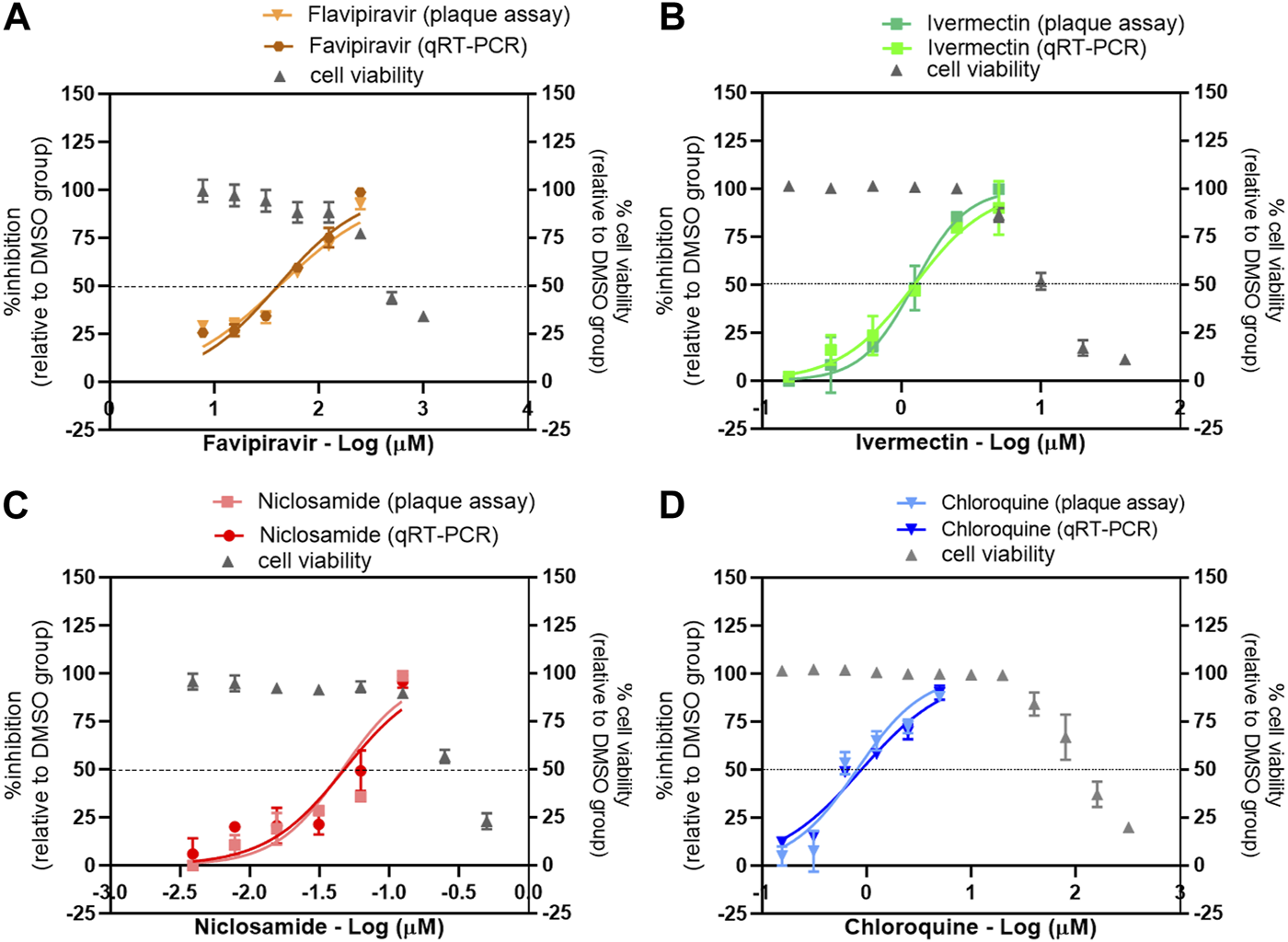
Evaluation of the antiviral activity of drug candidates against SARS-CoV-2 in vitro. The dose-response curves of a single drug treatment against SARS-CoV-2 are shown: (A) favipiravir, (B) ivermectin, (C) niclosamide, and (D) chloroquine. Vero E6 cells were pretreated with twofold serial dilutions of the drug for 1 h and infected with SARS-CoV-2 at an MOI of 0.01. The infected cells were maintained in the medium containing serial dilutions of the single drugs or 0.5% DMSO for 2 days. The virus production was determined using the plaque assay and one-step qRT-PCR. The dose-response curves are expressed as the percent inhibition relative to the DMSO-treated cell control. The effect of drug treatment on cell viability was determined using a MTT assay and expressed relative to the DMSO-treated cell control.
Two-drug combination treatments against SARS-CoV-2 in Vero E6 cells
The antiviral activities of favipiravir and the repurposed anti-parasitic drug combinations were assessed in vitro to find the combinations with good potential for COVID-19 treatment. Two-drug combinations were evaluated in Vero E6 cells, including favipiravir-ivermectin, favipiravir-niclosamide, and favipiravir-chloroquine. The evaluations of two-drug combinations were performed following the same infection protocol by treating with 16 different pairwise combinations of two drugs. All 16 pairwise two-drug combinations showed no significant cytotoxicity.
Favipiravir-ivermectin combination
The presence of ivermectin at various concentrations ranging from 0.3 to 2.4 µM induced a strong shift in the dose-response curves of favipiravir (Figure 2A). However, the accurate IC50 values of the dose-response curves with minimal antiviral activity above 50% could not be determined. Thus, the extrapolated IC50 values and fold reduction were dismissed (Table 2). In a similar manner, the presence of favipiravir also induced a strong shift in ivermectin dose-response curves with the presence of 80, 40, and 20 µM of favipiravir, while the presence of 10 µM of favipiravir resulted in a 4.28-fold reduction of the ivermectin IC50 value (Figure 2B; Table 2). All pairwise combinations showed increasing inhibitory effects compared to single drug treatments, with a maximum antiviral activity of approximately 90% (Figure 2C). The mean Loewe synergy scores of 13.43 accounted for the synergistic effect between favipiravir and ivermectin, with peak scores of 27.28 and 95% confidence intervals (CI) [21.56, 32.71] (Figure 2D). Moreover, the synergy score calculation using the HSA reference model also showed synergistic effects, with a mean synergy score of 23.79. However, the use of ZIP and Bliss independence models gave values of 9.94 and 9.88, respectively, which indicated the additive effect between favipiravir and ivermectin.
FIGURE 2
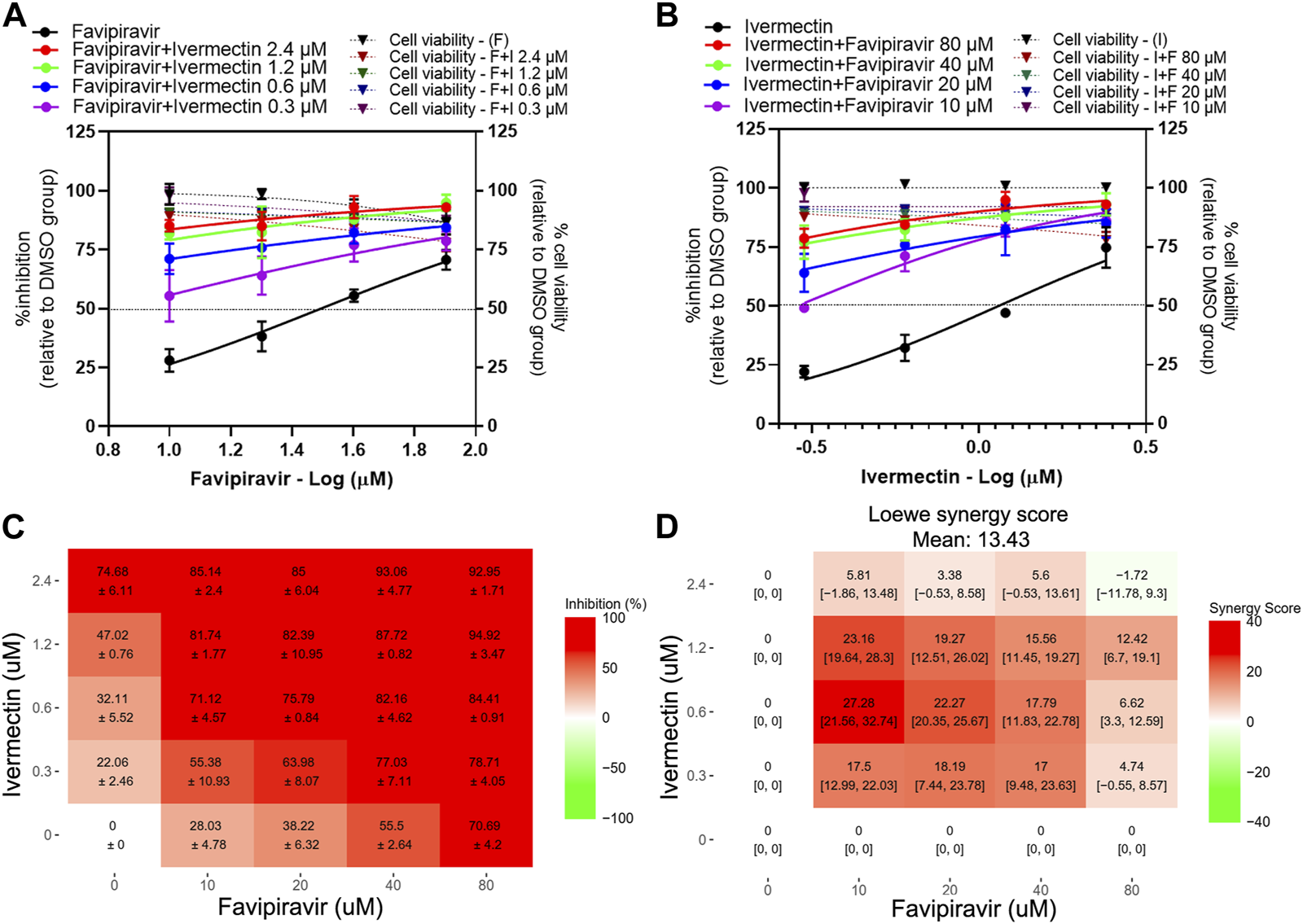
Evaluation of favipiravir-ivermectin combination treatments against SARS-CoV-2 in Vero E6 cells. Vero E6 cells were treated for 1 h with twofold serial dilutions of favipiravir in the presence of different fixed concentrations of ivermectin (A) or, alternatively, serial dilutions of ivermectin in the presence of different fixed concentrations of favipiravir (B). The cells were infected with SARS-CoV-2 at MOI 0.01. After removing the virus inoculum, the cells were further maintained in the medium containing drugs for 2 days. Virus production was determined using one-step qRT-PCR. The dose-response matrix of a two-drug combination is shown in (C). The synergy scores of two-drug combinations (D) were calculated using SynergyFinderPlus. For synergy scores less than −10, the interaction between two drugs was antagonistic. On scores from −10 to 10, the interaction between two drugs accounted for additive effects. For scores larger than 10, the interaction between two drugs accounted for the synergistic effect. The independent experiments were performed in triplicate, and the data are shown as mean ± SD in (A–C) or mean [95% confidence intervals (CI)] in (D) (Abbreviation: F, favipiravir; I, ivermectin).
TABLE 2
| Drug treatment | IC50 (µM) qRT-PCR | Fold reduction of IC50 (single/combined) | |
|---|---|---|---|
| Favipiravir-ivermectin | Favipiravir | 40.49 | |
| Favipiravir + ivermectin 2.4 µM | ND | ND | |
| Favipiravir + ivermectin 1.2 µM | ND | ND | |
| Favipiravir + ivermectin 0.6 µM | ND | ND | |
| Favipiravir + ivermectin 0.3 µM | ND | ND | |
| Ivermectin | 1.24 | ||
| Ivermectin + favipiravir 80 µM | ND | ND | |
| Ivermectin + favipiravir 40 µM | ND | ND | |
| Ivermectin + favipiravir 20 µM | ND | ND | |
| Ivermectin + favipiravir 10 µM | 0.29 | 4.28 | |
| Favipiravir-niclosamide | Favipiravir | 40.49 | |
| Favipiravir + niclosamide 0.09 µM | ND | ND | |
| Favipiravir + niclosamide 0.045 µM | ND | ND | |
| Favipiravir + niclosamide 0.0225 µM | 8.51 | 4.76 | |
| Favipiravir + niclosamide 0.01125 µM | 7.86 | 5.15 | |
| Niclosamide | 0.048 | ||
| Niclosamide + favipiravir 80 µM | ND | ND | |
| Niclosamide + favipiravir 40 µM | ND | ND | |
| Niclosamide + favipiravir 20 µM | ND | ND | |
| Niclosamide + favipiravir 10 µM | 0.014 | 3.428 | |
| Favipiravir-chloroquine | Favipiravir | 40.49 | |
| Favipiravir + chloroquine 1.7 µM | ND | ND | |
| Favipiravir + chloroquine 0.85 µM | ND | ND | |
| Favipiravir + chloroquine 0.425 µM | 13.01 | 3.11 | |
| Favipiravir + chloroquine 0.2125 µM | 23.48 | 1.72 | |
| Chloroquine | 0.89 | ||
| Chloroquine + favipiravir 80 µM | ND | ND | |
| Chloroquine + favipiravir 40 µM | ND | ND | |
| Chloroquine + favipiravir 20 µM | ND | ND | |
| Chloroquine + favipiravir 10 µM | 0.60 | 1.48 | |
Antiviral activity of two-drug combination treatments against SARS-CoV-2 in Vero E6 cells.
ND, cannot determine the IC50 values; a dose-response curve for a treatment using a two-drug combination showed an inhibition level greater than 50% at these concentrations.
Favipiravir-niclosamide combination
The presence of 0.09 and 0.045 µM niclosamide induced a strong shift in the dose-response curves of favipiravir, with the inhibitory activity ranging from 80% to 98% (Figures 3A, C). The presence of 0.0255 and 0.01125 µM niclosamide resulted in a 4.76- and 5.15-fold reduction of favipiravir IC50 values, respectively (Figure 3A; Table 2). Similarly, the presence of 80, 40, and 20 µM favipiravir also induced a strong shift in the niclosamide dose-response curves. While the presence of 10 µM of favipiravir resulted in a 3.428-fold reduction of the niclosamide IC50 value (Figure 3B; Table 2). The dose-response matrix shows an increasing inhibitory effect due to the increasing concentrations of favipiravir and niclosamide (Figure 3C). The mean Loewe synergy scores of 7.48 accounted for the additive effect between favipiravir and niclosamide, with peak scores of 16.86, 95% CI [12.88, 23.64] (Figure 3D). The synergy scores of 4.29 and 4.37 were obtained when using the ZIP and Bliss independence reference models, which also accounted for the additive effect between favipiravir and niclosamide. However, the synergy score calculation using the ZIP reference model resulted in synergy scores of 16.27, which showed the synergistic effect of this combination.
FIGURE 3
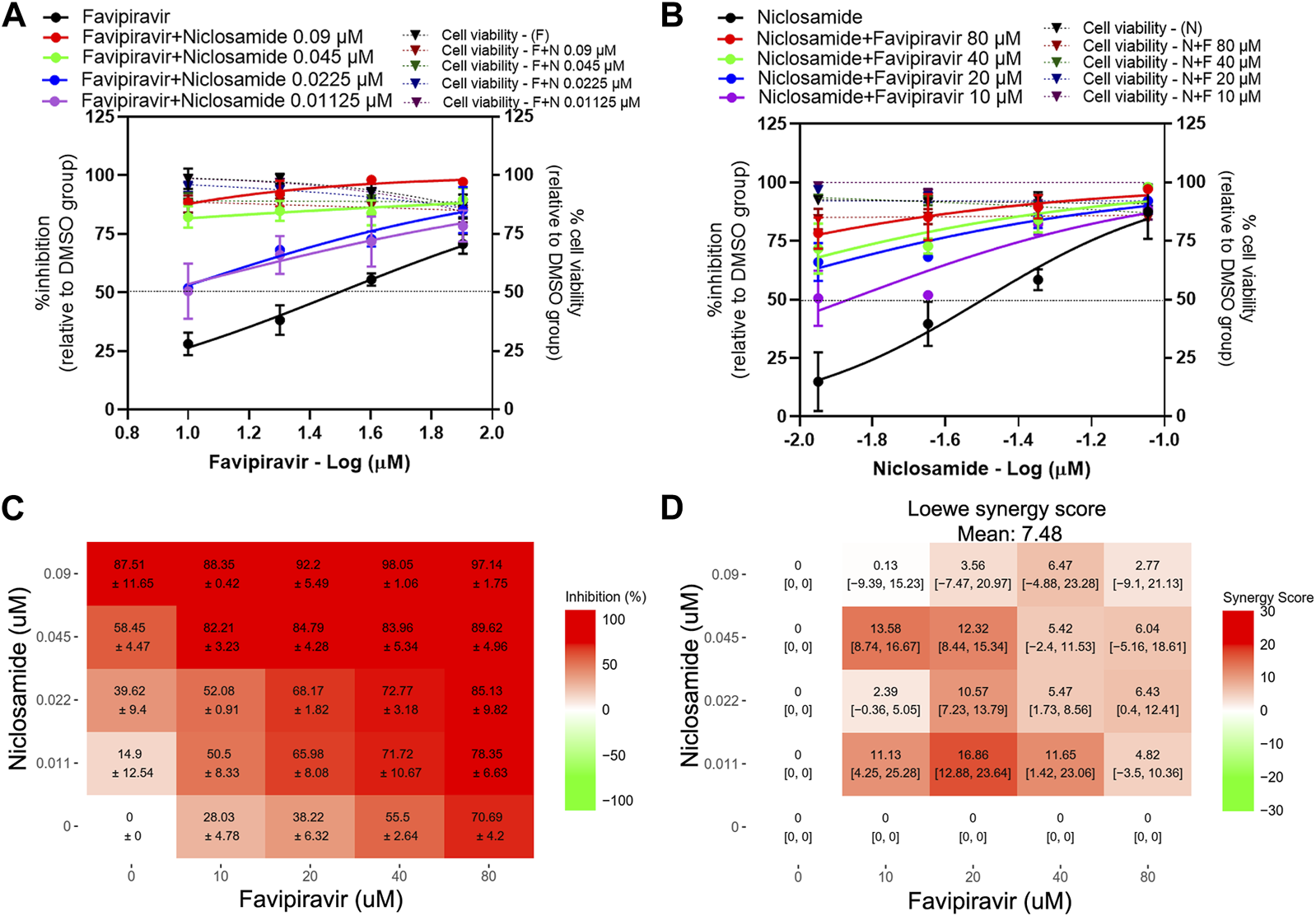
Evaluation of favipiravir-niclosamide combination treatments against SARS-CoV-2 in Vero E6 cells Vero E6 cells were treated for 1 h with twofold serial dilutions of favipiravir in the presence of different fixed concentrations of niclosamide (A) or, alternatively, serial dilutions of niclosamide in the presence of different fixed concentrations of favipiravir (B). The cells were infected with SARS-CoV-2 at MOI 0.01. After removing the virus inoculum, the cells were further maintained in the medium containing drugs for 2 days. Virus production was determined using one-step qRT-PCR. The dose-response matrix of a two-drug combination is shown in (C). The synergy scores of two-drug combinations (D) were calculated using SynergyFinderPlus. For synergy scores less than −10, the interaction between two drugs was antagonistic. On scores from −10 to 10, the interaction between two drugs accounted for additive effects. For scores larger than 10, the interaction between two drugs accounted for the synergistic effect. The independent experiments were performed in triplicate, and the data are shown as mean ± SD in (A–C) or mean [95% confidence intervals (CI)] in (D) (Abbreviation: F, favipiravir; N, niclosamide).
Favipiravir-chloroquine combination
The results show that the presence of chloroquine induced a shift in the dose-response curves of favipiravir, with a 3.11 and 1.72-fold reduction of favipiravir IC50 values in the presence of 0.425 and 0.2125 µM chloroquine, respectively (Figure 4A; Table 2). The presence of favipiravir also induced a shift in the chloroquine dose-response curves, with a 1.48-fold reduction of the chloroquine IC50 value in the presence of 10 µM of favipiravir (Figure 4B; Table 2). The dose-response matrix shows the increasing inhibitory effect in pairwise combinations with high concentrations of favipiravir and chloroquine (Figure 4C). However, most parts of the synergy score map show negative to low positive scores, even with the high concentrations of both drugs (Figure 4D). The mean Loewe synergy scores of −0.37 accounted for the additive effect between favipiravir and chloroquine, with peak scores of 11.33, 95% CI [9.11, 14.15] in the combination of 40 µM favipiravir and 0.425 µM chloroquine. The synergy scores of −7.19, −7.62, and 7.79 were obtained when using ZIP, Bliss independence, and HSA reference models, which also accounted for the additive effect of favipiravir and chloroquine.
FIGURE 4
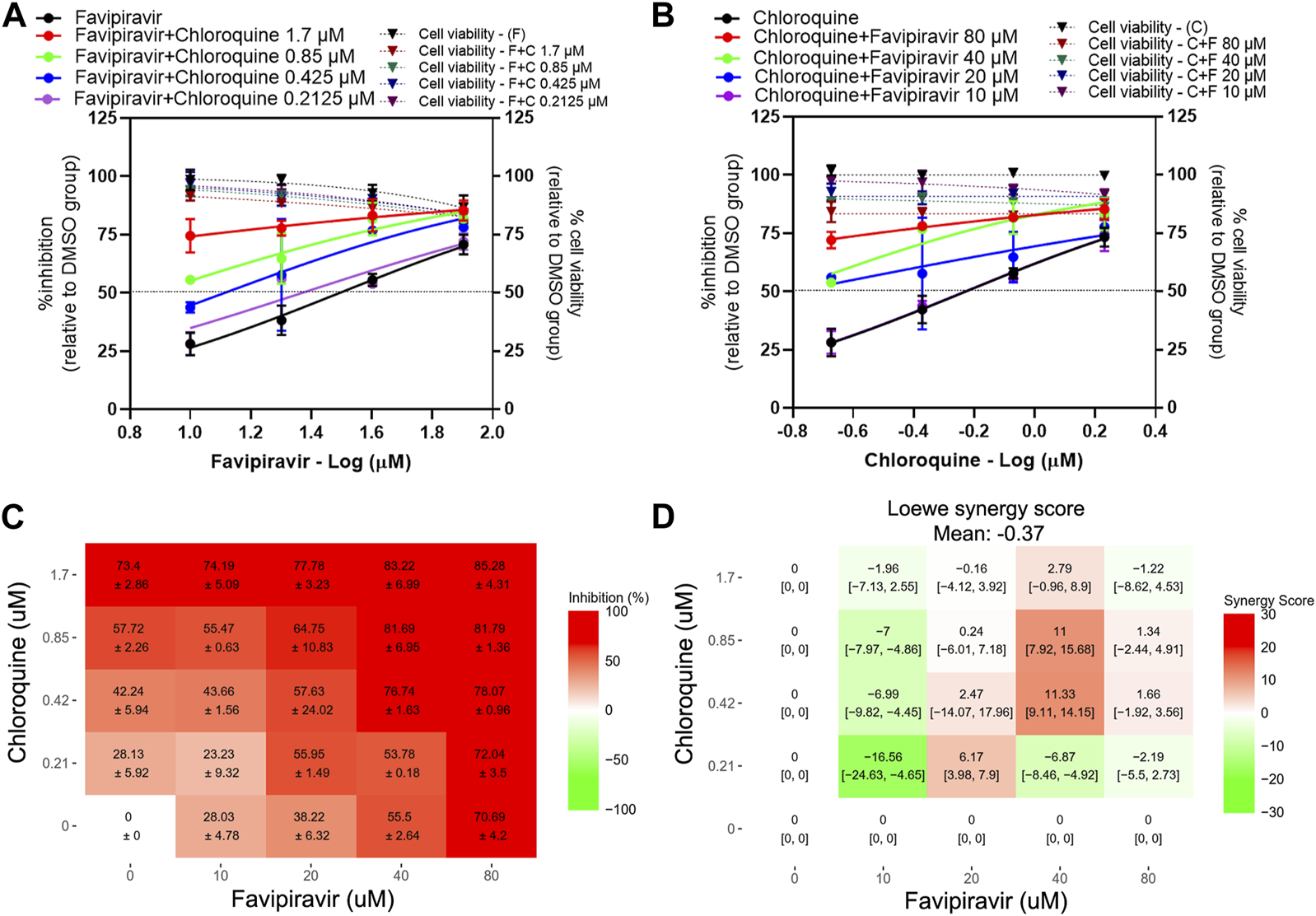
Evaluation of favipiravir-chloroquine combination treatments against SARS-CoV-2 in Vero E6 cells. Vero E6 cells were treated for 1 h with twofold serial dilutions of favipiravir in the presence of different fixed concentrations of chloroquine (A) or, alternatively, serial dilutions of chloroquine in the presence of different fixed concentrations of favipiravir (B). The cells were infected with SARS-CoV-2 at MOI 0.01. After removing the virus inoculum, the cells were further maintained in the medium containing drugs for 2 days. Virus production was determined using one-step qRT-PCR. The dose-response matrix of a two-drug combination is shown in (C). The synergy scores of two-drug combinations (D) were calculated using SynergyFinderPlus. For synergy scores less than −10, the interaction between two drugs was antagonistic. On scores from −10 to 10, the interaction between two drugs accounted for additive effects. For scores larger than 10, the interaction between two drugs accounted for the synergistic effect. The independent experiments were performed in triplicate, and the data are shown as mean ± SD in (A–C) or mean [95% confidence intervals (CI)] in (D) (Abbreviation: F, favipiravir; C, chloroquine).
Evaluation of single-drug treatment against SARS-CoV-2 in Calu-3 cells
Favipiravir-ivermectin combinations in Vero E6 cells demonstrated the synergistic effect. Thus, this combination was selected for further study in the human lung cancer cell line, Calu-3. Firstly, the inhibitory effects of single favipiravir and ivermectin treatments were determined using a plaque assay (Figure 5). The IC50 values of favipiravir and ivermectin were 913 µM and 0.2 µM, respectively. The effect of drug treatment on cell viability was determined using an MTT assay. The CC50 values of favipiravir and ivermectin were >2 mM and 3.09 µM. Therefore, the SI values of favipiravir and ivermectin were >2.19 and 15.45, respectively.
FIGURE 5
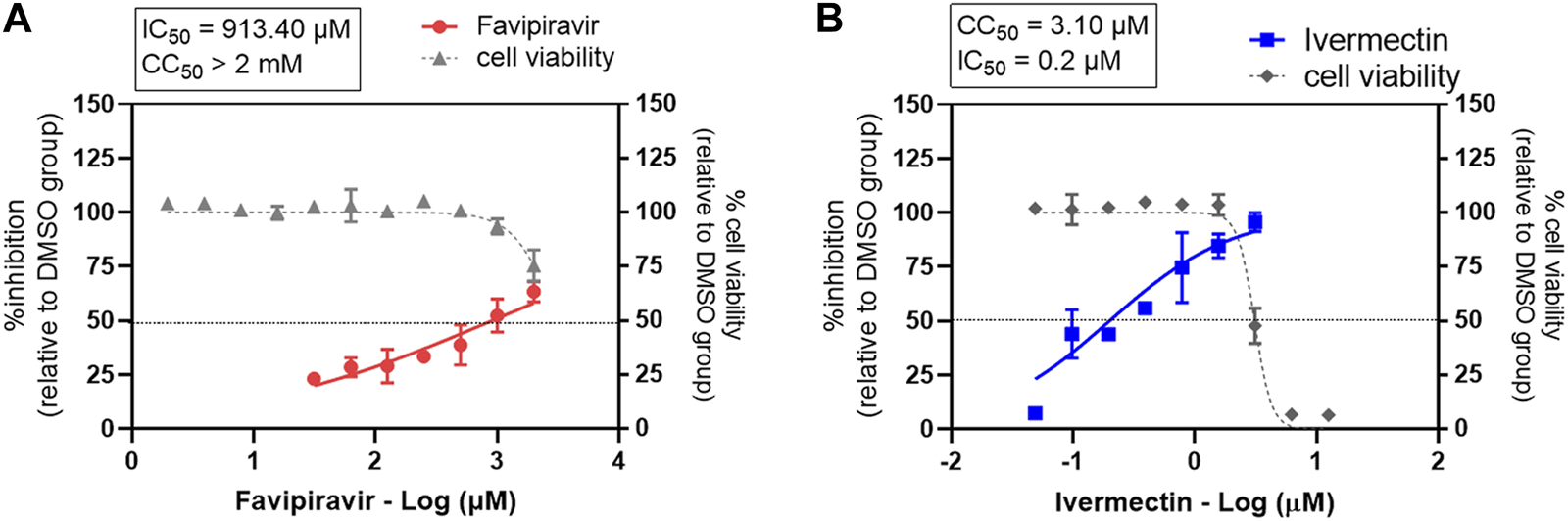
Evaluation of single-drug treatment against SARS-CoV-2 in Calu-3 cells. The dose-response curves of a single drug treatment against SARS-CoV-2 are shown for (A) favipiravir and (B) ivermectin. Calu-3 cells were treated with twofold serial dilutions of the drug for 1 h and infected with SARS-CoV-2. The infected cells were maintained in the medium containing serial dilutions of the single drug, or 0.5% DMSO, for 2 days. Virus production was determined using a plaque assay. The dose-response curves are expressed as percent inhibitions relative to the DMSO-treated cell control. The effect of drug treatment on cell viability was determined using a MTT assay and expressed relative to the DMSO-treated cell control. The experiments were performed in triplicate for drug treatments and in duplicate for cell viability assays; the data are shown in mean ± SD.
Evaluation of favipiravir-ivermectin combination treatment against SARS-CoV-2 in Calu-3 cells
The presence of 0.4 and 0.2 µM ivermectin induced a strong shift in the dose-response curves of favipiravir to a comparable level in every pairwise combination, ranging from 70% to 80% of inhibitory activity (Figure 6A). While the presence of 0.1 and 0.05 µM ivermectin induced a shift in the dose-response curve of favipiravir, with a 1.56 and 1.37-fold reduction of favipiravir IC50 values, respectively (Figure 6A; Table 3). Similarly, the presence of 1.8 and 0.9 mM favipiravir also induced a strong shift in the dose-response curves of ivermectin. The shift of ivermectin dose-response curves was observed with a 2.06 and 1.33-fold reduction of ivermectin IC50 values in the presence of 0.45 and 0.225 mM favipiravir, respectively (Figure 6B; Table 3). The dose-response matrix shows the increasing inhibitory effect in relation to the increasing drug concentrations (Figure 6C). The mean Loewe synergy scores of 1.44 accounted for the additive effect between favipiravir and ivermectin, with peak scores of 10.08, 95% CI [6.62, 15.30] (Figure 6D). Moreover, the synergy score calculation using ZIP, Bliss independence, and HSA reference models gave values of −2.21, −2.26, and 9.34, respectively, which indicated the additive effect between favipiravir and ivermectin. All 16 pairwise two-drug combinations showed no significant cytotoxicity (Figures 6A,B).
FIGURE 6
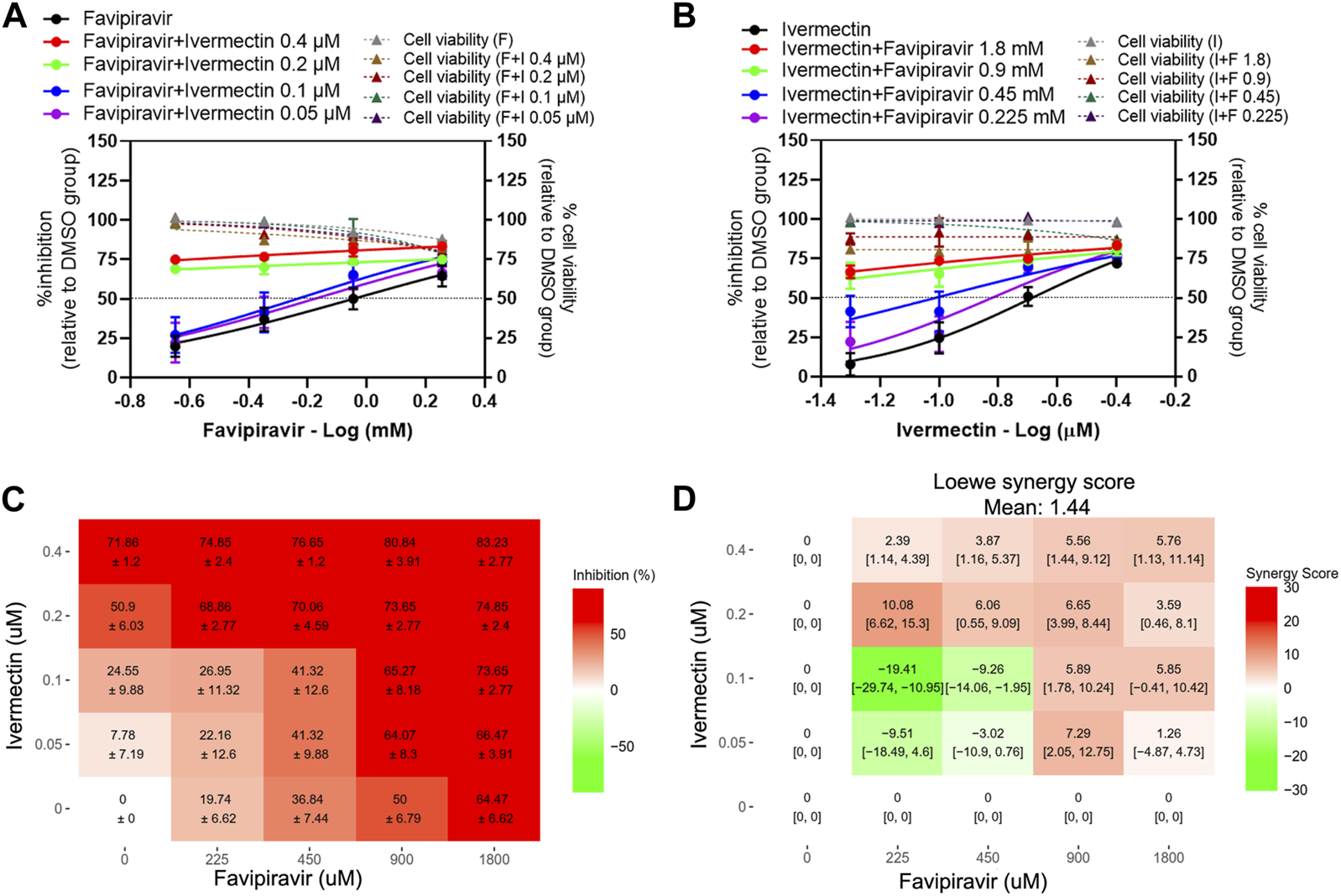
Evaluation of favipiravir-ivermectin combination treatments against SARS-CoV-2 in Calu-3 cells. Calu-3 cells were treated for 1 h with twofold serial dilutions of favipiravir in the presence of different fixed concentrations of ivermectin (A) or, alternatively, serial dilutions of ivermectin in the presence of different fixed concentrations of favipiravir (B). The cells were infected with SARS-CoV-2 at MOI 0.01. After removing the virus inoculum, the cells were further maintained in the medium containing drugs for 2 days. The virus production was determined using a plaque assay. The dose-response matrix of a two-drug combination is shown in (C). The synergy scores of two-drug combinations (D) were calculated using SynergyFinderPlus. For synergy scores less than −10, the interaction between two drugs was antagonistic. On scores from −10 to 10, the interaction between two drugs accounted for additive effects. For scores larger than 10, the interaction between two drugs accounted for the synergistic effect. The independent experiments were performed in triplicate, and the data are shown as mean ± SD in (A–C) or mean [95% confidence intervals (CI)] in (D) (Abbreviation: F, favipiravir; I, ivermectin).
TABLE 3
| Drug treatment | IC50 (µM) | Fold reduction of IC50 (single/combined) | |
|---|---|---|---|
| Favipiravir-Ivermectin | Favipiravir | 913 | |
| Favipiravir + ivermectin 0.4 µM | ND | ND | |
| Favipiravir + ivermectin 0.2 µM | ND | ND | |
| Favipiravir + ivermectin 0.1 µM | 583.5 | 1.56 | |
| Favipiravir + ivermectin 0.05 µM | 667.9 | 1.37 | |
| Ivermectin | 0.2 | ||
| Ivermectin + favipiravir 1.8 mM | ND | ND | |
| Ivermectin + favipiravir 0.9 mM | ND | ND | |
| Ivermectin + favipiravir 0.45 mM | 0.097 | 2.06 | |
| Ivermectin + favipiravir 0.225 mM | 0.151 | 1.33 | |
Favipiravir-ivermectin combination treatments against SARS-CoV-2 in Calu-3 cells.
ND, cannot determine the IC50 values; a dose-response curve for a treatment using a two-drug combination showed an inhibition level greater than 50% at these concentrations.
Discussion
Our results showed anti-SARS-CoV-2 activity of favipiravir in a high micromolar range (Vero E6: IC50 = 40.49 µM; Calu-3: IC50 = 913.4 µM), which is in line with the previous studies (Jeon et al., 2020; Shannon et al., 2020; Wang et al., 2020; Ohashi et al., 2021). The average maximum free plasma concentration of favipiravir ranges from 362 μM to 530 µM (Mentré et al., 2015; Nguyen et al., 2017; Hassanipour et al., 2021), which exceeds the drug IC50 value determined only using our Vero E6 model. A previous study demonstrated differences in favipiravir-RTP levels in different cell lines; higher favipiravir-RTP levels were observed in Vero, A549, and HEK293T compared to MDCK cells (Huchting et al., 2019). Further studies are required to assess the favipiravir-RTP levels and the activities of cellular enzymes required for phosphoribosylation in Calu-3 cells and lung cells.
The results of clinical trials demonstrated that favipiravir provided benefit to mild or moderate COVID-19-infected patients when taken early (Agrawal et al., 2020; Cai et al., 2020). However, the plasma concentration of favipiravir in critical patients was much lower, ranging from 28.2 µM to lower than the limit of quantification (>1 μg/mL or 6.37 µM) (Favié et al., 2018; Irie et al., 2020), which does not reach the IC50 concentration, thereby raising a concern in severe COVOD-19 treatment. Notably, the in vivo studies in a hamster model showed that the anti-SARS-CoV-2 activity of favipiravir could be achieved with a very high dose (Kaptein et al., 2020; Driouich et al., 2021). Therefore, a drug combination approach should be used to improve the antiviral activity of favipiravir. In this study, repurposed anti-parasitic drugs with broad-spectrum antiviral activity were selected for the two-drug combination treatment, including ivermectin, niclosamide, and chloroquine.
Ivermectin showed potent in vitro anti-SARS-CoV-2 activity in the low micromolar range (Vero E6: IC50 = 1.24 µM; Calu-3: IC50 = 0.2 µM). However, many clinical trials showed varying results in COVID-19 treatment, ranging from efficacious to no significant benefit (Vallejos et al., 2020; Bryant et al., 2021; Popp et al., 2021). The maximum plasma concentration of ivermectin was 0.059 µM, which was below the IC50 value for anti-SARS-CoV-2 activity (Caly et al., 2020; Schmith et al., 2020). However, higher concentrations of ivermectin were detected in various tissues, including fat, skin, and nodular tissues (Baraka et al., 1996), suggesting the potential of ivermectin in antiviral therapy. Additionally, a clinical study reported that ivermectin was safe and reduced the level of plasma nonstructural protein 1 in dengue patients (Suputtamongkol et al., 2021).
To improve the antiviral activity of these drugs, a drug combination approach was used. Favipiravir and ivermectin showed in vitro synergistic antiviral activity against SARS-CoV-2 in Vero E6 cells. The presence of ivermectin lowered the favipiravir IC50 value. A similar trend was observed for favipiravir, which also lowered the ivermectin IC50 value below the maximum plasma concentration. However, in Calu-3 cells, the favipiravir-ivermectin combination only showed an additive effect. This suggested that a drug combination strategy would help improve antiviral activity. Additionally, the combination of remdesivir and ivermectin also showed synergistic effects (Jeffreys et al., 2020; Tan et al., 2021). As both remdesivir and favipiravir target viral RdRp, their interaction with ivermectin may represent a similar synergistic mechanism.
Niclosamide also showed potent in vitro anti-SARS-CoV-2 activity in a low micromolar range (Vero E6: IC50 = 0.048 µM). The favipiravir-niclosamide combination demonstrated only an additive effect in Vero E6 cells. Similarly, the drug combination lowered the IC50 values of both favipiravir and niclosamide. The in vitro anti-SARS-CoV-2 activity of chloroquine showed a low micromolar range in Vero E6 cells. However, a recent study reported that chloroquine could efficiently block virus infection in Vero and Huh7.5 cells but not in Calu-3 cells (Hoffmann et al., 2020b). Vero and Huh7.5 cells require endosomal acidification during viral entry, while the entry in Calu-3 cell is pH-independent by using TMPRSS2, a transmembrane serine protease, to cleave the Spike protein extracellularly and bypass the pH-dependent step (Hoffmann et al., 2020a; Dittmar et al., 2021). Moreover, in vivo and clinical studies showed that chloroquine treatment in COVID-19 patients was ineffective (Kashour et al., 2021). In this study, the favipiravir-choroquine combination only demonstrated a minimum additive effect. It was previously shown by mathematical modeling that drugs working at the entry step performed poorly in synergy with other drugs (Melville et al., 2018). The lack of good synergy with chloroquine supports this model. A previous study demonstrated an antagonistic effect of the hydroxychloroquine and remdesivir combination against SARS-CoV-2, while nitazoxanide (which should be similar to niclosamide) made good synergy with various drugs (Bobrowski et al., 2021).
The drug combination approach maximizes the antiviral activity with a reduced dosage of drugs needed, thus minimizing the side effects and the risk of resistance development. In particular, the combination of antiviral drugs with drugs utilizing different modes of action or acting on cellular targets helps reduce cellular toxicity and drug resistance. (Day and Siu, 2016). There are several successful drug combinations used in the treatment of HIV-1 and the hepatitis C virus (WHO, 2018; AASLD, 2021). Moreover, the combination of favipiravir and ivermectin is being tested in a randomized controlled clinical trial (ClinicalTrials.gov Identifier: NCT05155527).
In summary, our study demonstrated that the antiviral effects of single drugs were only achieved by using high doses. The drug combination strategy is able to reduce the IC50 values of individual drugs, providing the treatment with more options. Thus, the combinations of favipiravir and ivermectin or niclosamide are the attractive drug combinations to test in clinical trials due to their synergism, relatively good safety profile, and wide availability.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
KJ: Methodology, Formal Analysis, Investigation, Writing–Original Draft Preparation. CB: Methodology, Formal Analysis, Investigation. SM: Methodology, Investigation. NP: Methodology, Investigation. SB: Resource. AT: Validation, Methodology. PaA: Validation, Methodology. PrA: Conceptualization, Supervision, Funding, Writing–Review and Editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Chair Professor Program (P-20-52262), The National Science and Technology Development Agency (NSTDA), Thailand. This study was partly supported by Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand. The funders had no role in study design, data collection, analysis, and interpretation, or in writing the manuscript.
Acknowledgments
We are grateful to the members of Siriraj Center of Research Excellence in Dengue and Emerging Pathogens, and AT’s laboratory for all support and assistance provided throughout the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2023.12265/full#supplementary-material
Abbreviations
Bliss, Bliss independence; CC50, 50% cytotoxic concentration; COVID-19, corona virus disease of 2019; CPE, cytopathic effect; Ct, threshold cycle; DMEM/F12, Dulbecco’s Modified Eagle Medium/Nutrient Mixture F12; DMSO, dimethyl sulfoxide; EC50, half maximum effective concentration; favipiravir-RTP, favipiravir-ribofuranosyl-5′-triphosphate; FDA, Food and drug administration; FBS, fetal bovine serum; HSA, Highest Single Agent; HIV-1, human immunodeficiency virus type 1; IC50, half-maximal inhibitory concentration; Loewe, Loewe additivity; MEM, Minimum Essential Medium; MOI, multiplicity of infection; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; RdRp, RNA-dependent RNA polymerase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SI, selectivity index; TCID50, 50% cell culture infectious dose; Tm, melting temperature; WHO, World Health Organization; ZIP, Zero Independence Potency.
References
1
AASLD (2021). HCV Guidance: recommendations for testing, managing, and treating Hepatitis C Virus infection. United States: Infectious Diseases Society of America.
2
Agrawal U. Raju R. Udwadia Z. F. (2020). Favipiravir: a new and emerging antiviral option in COVID-19. Med. J. Armed Forces India76, 370–376. 10.1016/j.mjafi.2020.08.004
3
Ahmed S. Karim M. M. Ross A. G. Hossain M. S. Clemens J. D. Sumiya M. K. et al (2021). A five-day course of ivermectin for the treatment of COVID-19 may reduce the duration of illness. Int. J. Infect. Dis.103, 214–216. 10.1016/j.ijid.2020.11.191
4
Alam S. Kamal T. B. Sarker M. M. R. Zhou J. R. Rahman S. M. A. Mohamed I. N. (2021). Therapeutic effectiveness and safety of repurposing drugs for the treatment of COVID-19: Position standing in 2021. Front. Pharmacol.12, 659577. 10.3389/fphar.2021.659577
5
Ansems K. Grundeis F. Dahms K. Mikolajewska A. Thieme V. Piechotta V. et al (2021). Remdesivir for the treatment of COVID-19. Cochrane Database Syst. Rev.8, Cd014962. 10.1002/14651858.cd014962
6
Axfors C. Schmitt A. M. Janiaud P. van’t Hooft J. Abd-Elsalam S. Abdo E. F. et al (2021). Mortality outcomes with hydroxychloroquine and chloroquine in COVID-19 from an international collaborative meta-analysis of randomized trials. Nat. Commun.12, 2349. 10.1038/s41467-021-22446-z
7
Backer V. Sjöbring U. Sonne J. Weiss A. Hostrup M. Johansen H. K. et al (2021). A randomized, double-blind, placebo-controlled phase 1 trial of inhaled and intranasal niclosamide: a broad spectrum antiviral candidate for treatment of COVID-19. Lancet Reg. Health Eur.4, 100084. 10.1016/j.lanepe.2021.100084
8
Baraka O. Z. Mahmoud B. M. Marschke C. K. Geary T. G. Homeida M. M. Williams J. F. (1996). Ivermectin distribution in the plasma and tissues of patients infected with onchocerca volvulus. Eur. J. Clin. Pharmacol.50, 407–410. 10.1007/s002280050131
9
Baranovich T. Wong S. S. Armstrong J. Marjuki H. Webby R. J. Webster R. G. et al (2013). T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol.87, 3741–3751. 10.1128/jvi.02346-12
10
Behera P. Patro B. K. Singh A. K. Chandanshive P. D. Pradhan S. K. Pentapati S. S. K. et al (2021). Role of ivermectin in the prevention of SARS-CoV-2 infection among healthcare workers in india: a matched case-control study. PLOS ONE16, e0247163. 10.1371/journal.pone.0247163
11
Bobrowski T. Chen L. Eastman R. T. Itkin Z. Shinn P. Chen C. Z. et al (2021). Synergistic and antagonistic drug combinations against SARS-CoV-2. Mol. Ther.29, 873–885. 10.1016/j.ymthe.2020.12.016
12
Bryant A. Lawrie T. A. Dowswell T. Fordham E. J. Mitchell S. Hill S. R. et al (2021). Ivermectin for prevention and treatment of COVID-19 infection: a systematic review, meta-analysis, and trial sequential analysis to inform clinical guidelines. Am. J. Ther.28, e434–e460. 10.1097/mjt.0000000000001402
13
Cai Q. Yang M. Liu D. Chen J. Shu D. Xia J. et al (2020). Experimental treatment with favipiravir for COVID-19: an open-label control study. Eng. (Beijing)6, 1192–1198. 10.1016/j.eng.2020.03.007
14
Caly L. Druce J. D. Catton M. G. Jans D. A. Wagstaff K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res.178, 104787. 10.1016/j.antiviral.2020.104787
15
Day D. Siu L. L. (2016). Approaches to modernize the combination drug development paradigm. Genome Med.8, 115. 10.1186/s13073-016-0369-x
16
Delang L. Abdelnabi R. Neyts J. (2018). Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antivir. Res.153, 85–94. 10.1016/j.antiviral.2018.03.003
17
Department of Medical Services (2020). COVID-19 in Thailand. Thailand: Department of Medical Services.
18
Dittmar M. Lee J. S. Whig K. Segrist E. Li M. Kamalia B. et al (2021). Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep.35, 108959. 10.1016/j.celrep.2021.108959
19
Driouich J.-S. Cochin M. Lingas G. Moureau G. Touret F. Petit P.-R. et al (2021). Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun.12, 1735. 10.1038/s41467-021-21992-w
20
Escribano-Romero E. Jiménez de Oya N. Domingo E. Saiz J. C. (2017). Extinction of west nile virus by favipiravir through lethal mutagenesis. Antimicrob. Agents Chemother.61, e01400-17. 10.1128/aac.01400-17
21
Fang J. Sun L. Peng G. Xu J. Zhou R. Cao S. et al (2013). Identification of three antiviral inhibitors against japanese encephalitis virus from library of pharmacologically active compounds 1280. PLOS ONE8, e78425. 10.1371/journal.pone.0078425
22
Favié L. M. Murk J. L. Meijer A. Nijstad A. L. van Maarseveen E. M. Sikma M. A. (2018). Pharmacokinetics of favipiravir during continuous venovenous haemofiltration in a critically ill patient with influenza. Antivir. Ther.23, 457–461. 10.3851/imp3210
23
Furuta Y. Komeno T. Nakamura T. (2017). Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci.93, 449–463. 10.2183/pjab.93.027
24
Ganguly D. Rottet S. Yee S. Hee W. Smith A. Khin N. et al (2020). SYBR green one-step qRT-PCR for the detection of SARS-CoV-2 RNA in saliva. BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2020.05.29.109702v2 (Accessed June 19, 2020).
25
Gassen N. C. Niemeyer D. Muth D. Corman V. M. Martinelli S. Gassen A. et al (2019). SKP2 attenuates autophagy through beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat. Commun.10, 5770. 10.1038/s41467-019-13659-4
26
Goldhill D. H. Yan A. Frise R. Zhou J. Shelley J. Gallego Cortés A. et al (2021). Favipiravir-resistant influenza A virus shows potential for transmission. PLoS Pathog.17, e1008937. 10.1371/journal.ppat.1008937
27
Götz V. Magar L. Dornfeld D. Giese S. Pohlmann A. Höper D. et al (2016). Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep.6, 23138. 10.1038/srep23138
28
Guedj J. Piorkowski G. Jacquot F. Madelain V. Nguyen T. H. T. Rodallec A. et al (2018). Antiviral efficacy of favipiravir against ebola virus: a translational study in cynomolgus macaques. PLoS Med.15, e1002535. 10.1371/journal.pmed.1002535
29
Guzzo C. A. Furtek C. I. Porras A. G. Chen C. Tipping R. Clineschmidt C. M. et al (2002). Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects. J. Clin. Pharmacol.42, 1122–1133. 10.1177/009127002237994
30
Hassanipour S. Arab-Zozani M. Amani B. Heidarzad F. Fathalipour M. Martinez-de-Hoyo R. (2021). The efficacy and safety of favipiravir in treatment of COVID-19: a systematic review and meta-analysis of clinical trials. Sci. Rep.11, 11022. 10.1038/s41598-021-90551-6
31
Hoffmann M. Kleine-Weber H. Schroeder S. Krüger N. Herrler T. Erichsen S. et al (2020a). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell181, 271–280.e8. 10.1016/j.cell.2020.02.052
32
Hoffmann M. Mösbauer K. Hofmann-Winkler H. Kaul A. Kleine-Weber H. Krüger N. et al (2020b). Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature585, 588–590. 10.1038/s41586-020-2575-3
33
Huang L. Yang M. Yuan Y. Li X. Kuang E. (2017). Niclosamide inhibits lytic replication of epstein-Barr virus by disrupting mTOR activation. Antivir. Res.138, 68–78. 10.1016/j.antiviral.2016.12.002
34
Huchting J. Vanderlinden E. Van Berwaer R. Meier C. Naesens L. (2019). Cell line-dependent activation and antiviral activity of T-1105, the non-fluorinated analogue of T-705 (favipiravir). Antivir. Res.167, 1–5. 10.1016/j.antiviral.2019.04.002
35
Hurt A. C. Wheatley A. K. (2021). Neutralizing antibody therapeutics for COVID-19. Viruses13, 628. 10.3390/v13040628
36
Ianevski A. Giri A. K. Aittokallio T. (2020). SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res.48, W488–W493. 10.1093/nar/gkaa216
37
Irie K. Nakagawa A. Fujita H. Tamura R. Eto M. Ikesue H. et al (2020). Pharmacokinetics of favipiravir in critically ill patients with COVID-19. Clin. Transl. Sci.13, 880–885. 10.1111/cts.12827
38
Jeffreys L. Pennington S. H. Duggan J. Breen A. Jinks J. Ardrey A. et al (2020). Remdesivir-Ivermectin combination displays synergistic interaction with improved in vitro antiviral activity against SARS-CoV-2. BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2020.12.23.424232v2 (Accessed September 01, 2021).
39
Jeon S. Ko M. Lee J. Choi I. Byun S. Y. Park S. et al (2020). Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother.64, e00819-20. 10.1128/aac.00819-20
40
Jitobaom K. Boonarkart C. Manopwisedjaroen S. Punyadee N. Borwornpinyo S. Thitithanyanont A. et al (2022). Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations. BMC Pharmacol. Toxicol.23, 41. 10.1186/s40360-022-00580-8
41
Jochmans D. van Nieuwkoop S. Smits S. L. Neyts J. Fouchier R. A. van den Hoogen B. G. (2016). Antiviral activity of Favipiravir (T-705) against a broad range of paramyxoviruses in vitro and against human metapneumovirus in hamsters. Antimicrob. Agents Chemother.60, 4620–4629. 10.1128/aac.00709-16
42
Kadri H. Lambourne O. A. Mehellou Y. (2018). Niclosamide, a drug with many repurposes. Chem. Med. Chem.13, 1088–1091. 10.1002/cmdc.201800100
43
Kamat S. Kumari M. (2021). Repurposing chloroquine against multiple diseases with special attention to SARS-CoV-2 and associated toxicity. Front. Pharmacol.12, 576093. 10.3389/fphar.2021.576093
44
Kanjanasirirat P. Suksatu A. Manopwisedjaroen S. Munyoo B. Tuchinda P. Jearawuttanakul K. et al (2020). High-content screening of thai medicinal plants reveals boesenbergia rotunda extract and its component panduratin A as anti-SARS-CoV-2 agents. Sci. Rep.10, 19963. 10.1038/s41598-020-77003-3
45
Kao J.-C. HuangFu W.-C. Tsai T.-T. Ho M.-R. Jhan M.-K. Shen T.-J. et al (2018). The antiparasitic drug niclosamide inhibits dengue virus infection by interfering with endosomal acidification independent of mTOR. PLoS neglected Trop. Dis.12, e0006715. 10.1371/journal.pntd.0006715
46
Kaptein S. J. F. Jacobs S. Langendries L. Seldeslachts L. Ter Horst S. Liesenborghs L. et al (2020). Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. U. S. A.117, 26955–26965. 10.1073/pnas.2014441117
47
Kashour Z. Riaz M. Garbati M. A. AlDosary O. Tlayjeh H. Gerberi D. et al (2021). Efficacy of chloroquine or hydroxychloroquine in COVID-19 patients: a systematic review and meta-analysis. J. Antimicrob. Chemother.76, 30–42. 10.1093/jac/dkaa403
48
Kim J. A. Seong R. K. Kumar M. Shin O. S. (2018). Favipiravir and ribavirin inhibit replication of asian and african strains of Zika virus in different cell models. Viruses10, 72. 10.3390/v10020072
49
Kongmanas K. Punyadee N. Wasuworawong K. Songjaeng A. Prommool T. Pewkliang Y. et al (2020). Immortalized stem cell-derived hepatocyte-like cells: an alternative model for studying dengue pathogenesis and therapy. PLoS neglected Trop. Dis.14, e0008835. 10.1371/journal.pntd.0008835
50
Kory P. Meduri G. U. Varon J. Iglesias J. Marik P. E. (2021). Review of the emerging evidence demonstrating the efficacy of ivermectin in the prophylaxis and treatment of COVID-19. Am. J. Ther.28, e299–e318. 10.1097/mjt.0000000000001377
51
Li Y. Li P. K. Roberts M. J. Arend R. C. Samant R. S. Buchsbaum D. J. (2014). Multi-targeted therapy of cancer by niclosamide: a new application for an old drug. Cancer Lett.349, 8–14. 10.1016/j.canlet.2014.04.003
52
Lim S. C. L. Hor C. P. Tay K. H. Mat Jelani A. Tan W. H. Ker H. B. et al (2022). Efficacy of ivermectin treatment on disease progression among adults with mild to moderate COVID-19 and comorbidities: the I-tech randomized clinical trial. JAMA Intern. Med.182, 426. 10.1001/jamainternmed.2022.0189
53
Lima-Morales R. Méndez-Hernández P. Flores Y. N. Osorno-Romero P. Cuecuecha-Rugerio E. Nava-Zamora A. et al (2021). Effectiveness of a multidrug therapy consisting of ivermectin, azithromycin, montelukast and acetylsalicylic acid to prevent hospitalization and death among ambulatory COVID-19 cases in tlaxcala, mexico. Int. J. Infect. Dis.105, 598–605. 10.1016/j.ijid.2021.02.014
54
Mazzon M. Ortega-Prieto A. M. Imrie D. Luft C. Hess L. Czieso S. et al (2019). Identification of broad-spectrum antiviral compounds by targeting viral entry. Viruses11, 176. 10.3390/v11020176
55
Melville K. Rodriguez T. Dobrovolny H. M. (2018). Investigating different mechanisms of action in combination therapy for influenza. Front. Pharmacol.9, 1207. 10.3389/fphar.2018.01207
56
Mentré F. Taburet A.-M. Guedj J. Anglaret X. Keïta S. de Lamballerie X. et al (2015). Dose regimen of favipiravir for ebola virus disease. Lancet. Infect. Dis.15, 150–151. 10.1016/s1473-3099(14)71047-3
57
Musa I. R. (2020). Potential antiviral effect of chloroquine therapy against SARS-CoV-2 infection. Open Access Maced. J. Med. Sci.8, 184–191. 10.3889/oamjms.2020.4854
58
Ngo B. T. Marik P. Kory P. Shapiro L. Thomadsen R. Iglesias J. et al (2021). The time to offer treatments for COVID-19. Expert Opin. Investig. Drugs30, 505–518. 10.1080/13543784.2021.1901883
59
Nguyen T. H. T. Guedj J. Anglaret X. Laouénan C. Madelain V. Taburet A.-M. et al (2017). Favipiravir pharmacokinetics in ebola-Infected patients of the JIKI trial reveals concentrations lower than targeted. PLoS neglected Trop. Dis.11, e0005389. 10.1371/journal.pntd.0005389
60
Niyomdecha N. Suptawiwat O. Boonarkart C. Jitobaom K. Auewarakul P. (2020). Inhibition of human immunodeficiency virus type 1 by niclosamide through mTORC1 inhibition. Heliyon6, e04050. 10.1016/j.heliyon.2020.e04050
61
Ohashi H. Watashi K. Saso W. Shionoya K. Iwanami S. Hirokawa T. et al (2021). Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience24, 102367. 10.1016/j.isci.2021.102367
62
Phougat N. Khatri S. Singh A. Dangi M. Kumar M. Dabur R. et al (2014). Combination therapy: the propitious rationale for drug development. Comb. Chem. High. Throughput Screen17, 53–67. 10.2174/13862073113166660065
63
Pires de Mello C. P. Tao X. Kim T. H. Vicchiarelli M. Bulitta J. B. Kaushik A. et al (2018). Clinical regimens of Favipiravir inhibit zika virus replication in the hollow-fiber infection model. Antimicrob. Agents Chemother.62, e00967-18. 10.1128/aac.00967-18
64
Popp M. Metzendorf M. I. Gould S. Kranke P. Meybohm P. Skoetz N. et al (2021). Ivermectin for preventing and treating COVID-19. Cochrane Database Syst. Rev.28, CD015017. 10.1002/14651858.CD015017.pub3
65
Reed L. J. Muench H. (1938). A simple method of estimating fifty percent enpoint. Am. J. Epidemiol.27, 493–497. 10.1093/oxfordjournals.aje.a118408
66
Schmith V. D. Zhou J. Lohmer L. R. L. (2020). The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacol. Ther.108, 762–765. 10.1002/cpt.1889
67
Shannon A. Selisko B. Le N.-T.-T. Huchting J. Touret F. Piorkowski G. et al (2020). Rapid incorporation of favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun.11, 4682. 10.1038/s41467-020-18463-z
68
Singh B. Ryan H. Kredo T. Chaplin M. Fletcher T. (2021). Chloroquine or hydroxychloroquine for prevention and treatment of COVID-19. Cochrane Database Syst. Rev.2, Cd013587. 10.1002/14651858.cd013587.pub2
69
Suputtamongkol Y. Avirutnan P. Mairiang D. Angkasekwinai N. Niwattayakul K. Yamasmith E. et al (2021). Ivermectin accelerates circulating nonstructural protein 1 (NS1) clearance in adult dengue patients: a combined phase 2/3 randomized double-blinded placebo controlled trial. Clin. Infect. Dis.72, e586–e593. 10.1093/cid/ciaa1332
70
Tan Y. L. Tan K. S. W. Chu J. J. H. Chow V. T. (2021). Combination treatment with remdesivir and ivermectin exerts highly synergistic and potent antiviral activity against murine coronavirus infection. Front. Cell. Infect. Microbiol.11, 700502. 10.3389/fcimb.2021.700502
71
US FDA (2021). Why you should not use ivermectin to treat or prevent COVID-19. US: FDA.
72
Vallejos J. Zoni R. Bangher M. Villamandos S. Bobadilla A. Plano F. et al (2020). Ivermectin to prevent hospitalizations in patients with COVID-19 (IVERCOR-COVID19): A structured summary of a study protocol for a randomized controlled trial. Trials21, 965. 10.1186/s13063-020-04813-1
73
Vallejos J. Zoni R. Bangher M. Villamandos S. Bobadilla A. Plano F. et al (2021). Ivermectin to prevent hospitalizations in patients with COVID-19 (IVERCOR-COVID19) a randomized, double-blind, placebo-controlled trial. BMC Infect. Dis.21, 635. 10.1186/s12879-021-06348-5
74
Vincent M. J. Bergeron E. Benjannet S. Erickson B. R. Rollin P. E. Ksiazek T. G. et al (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J.2, 69. 10.1186/1743-422x-2-69
75
Wagstaff K. M. Sivakumaran H. Heaton S. M. Harrich D. Jans D. A. (2012). Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J.443, 851–856. 10.1042/bj20120150
76
Wang M. Cao R. Zhang L. Yang X. Liu J. Xu M. et al (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res.30, 269–271. 10.1038/s41422-020-0282-0
77
Wang Y. Yuan C. Xu X. Chong T. H. Zhang L. Cheung P. P.-H. et al (2021b). The mechanism of action of T-705 as a unique delayed chain terminator on influenza viral polymerase transcription. Biophys. Chem.277, 106652. 10.1016/j.bpc.2021.106652
78
Wang Y.-Y. Huang Q. Shen Q. Zi H. Li B.-H. Li M.-Z. et al (2021a). Quality of and recommendations for relevant clinical practice guidelines for COVID-19 management: a systematic review and critical appraisal. Front. Med.8, 630765. 10.3389/fmed.2021.630765
79
Wen W. Chen C. Tang J. Wang C. Zhou M. Cheng Y. et al (2022). Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and paxlovid) for COVID-19: A meta-analysis. Ann. Med.54, 516–523. 10.1080/07853890.2022.2034936
80
WHO (2018). Updated recommendations on first-line and second-line antiretroviral regimens and post-exposure prophylaxis and recommendations on early infant diagnosis of HIV. København, Denmark: WHO.
81
WHO (2022). Therapeutics and COVID-19: living guideline. København, Denmark: WHO.
82
Worldometer (2022). COVID-19 coronavirus pandemic. Available at: https://www.worldometers.info/coronavirus/.
83
Xu J. Shi P.-Y. Li H. Zhou J. (2020). Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis.6, 909–915. 10.1021/acsinfecdis.0c00052
84
Xu T.-L. Han Y. Liu W. Pang X.-Y. Zheng B. Zhang Y. et al (2018). Antivirus effectiveness of ivermectin on dengue virus type 2 in aedes albopictus. PLoS neglected Trop. Dis.12, e0006934. 10.1371/journal.pntd.0006934
85
Yadav B. Wennerberg K. Aittokallio T. Tang J. (2015). Searching for drug synergy in complex dose–response landscapes using an interaction potency model. Comput. Struct. Biotechnol.13, 504–513. 10.1016/j.csbj.2015.09.001
86
Yang S. N. Y. Atkinson S. C. Wang C. Lee A. Bogoyevitch M. A. Borg N. A. et al (2020). The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antivir. Res.177, 104760. 10.1016/j.antiviral.2020.104760
87
Zheng S. Wang W. Aldahdooh J. Malyutina A. Shadbahr T. Tanoli Z. et al (2022). SynergyFinder plus: toward better interpretation and annotation of drug combination screening datasets. Genomics, Proteomics Bioinforma.20, 587–596. 10.1016/j.gpb.2022.01.004
Summary
Keywords
SARS-CoV-2, favipiravir, ivermectin, anti-parasitic drugs, repurposed drugs
Citation
Jitobaom K, Boonarkart C, Manopwisedjaroen S, Punyadee N, Borwornpinyo S, Thitithanyanont A, Avirutnan P and Auewarakul P (2023) Favipiravir and ivermectin show in vitro synergistic antiviral activity against SARS-CoV-2. Acta Virol. 67:12265. doi: 10.3389/av.2023.12265
Received
11 July 2022
Accepted
17 October 2023
Published
02 November 2023
Volume
67 - 2023
Edited by
Katarina Polcicova, Biomedical Research Center, Slovak Academy of Sciences, Slovakia
Updates
Copyright
© 2023 Jitobaom, Boonarkart, Manopwisedjaroen, Punyadee, Borwornpinyo, Thitithanyanont, Avirutnan and Auewarakul.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prasert Auewarakul, prasert.aue@mahidol.ac.th
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.