- 1Baotou Medical College of Inner Mongolia University of Science and Technology, Baotou, China
- 2Inner Mongolia Key Laboratory for Molecular Regulation of the Cell, College of Life Sciences, Inner Mongolia University, Hohhot, China
- 3Inner Mongolia Autonomous Region Center for Disease Control and Prevention, Hohhot, China
- 4Inner Mongolia Medical University, Hohhot, China
- 5Inner Mongolia Women and Children’s Hospital, Hohhot, China
Enterovirus 71 (EV71) is a significant causative agent of hand, foot and mouth disease (HFMD). However, the precise mechanism by which EV71 infection leads to alterations in the immune response remain elusive. To address this knowledge gap, we conducted a study where we introduced the Inner Mongolia EV71 C33λ strain into Vero cells, derived from African green monkey kidney cells. Subsequently, we performed RNA sequencing (RNA-Seq) to investigate the changes in the transcriptome of these infected Vero cells. Our primary objective was to establish a foundational understanding that could inform future research on EV71-associated immune factors. In our study, we identified a total of 942 differentially expressed genes (DEGs) in Vero cells infected with Enterovirus 71 (EV71), with 568 gene exhibiting increased expression and 374 gene showing decreased expression. To elucidate the functional implications of these DEGs, we conducted a comprehensive functional enrichment analysis using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. This analysis revealed three genes that were significantly upregulated, which we subsequently validated using reverse transcription polymerase chain reaction technology (RT-qPCR). The RT-qPCR results were in strong agreement with our RNA-Seq data, confirming the reliability of our findings. This study represents the pioneering RNA-Seq analysis that delves into the cellular response of Vero cells to EV71 infection. Our results not only provide a foundational understanding of the molecular changes induced by EV71 but also offer crucial insights into the mechanisms by which EV71 modulates the host immune system. These insights are pivotal for future research endeavors aimed at developing effective therapeutic strategies against EV71 and related pathogens, as well as for understanding the broader implications of viral infections on host immunity.
Introduction
Enterovirus 71 (EV71) is a single-stranded RNA-based pathogen from the Picornaviridae family (Liu et al., 2011), and is a non-enveloped virus with a 7.5 kb genome that contains an open reading frame (ORF) flanked by a 5′ untranslated region (5′UTR) and a polyadenylated 3′UTR (Oprisan et al., 2002; McMinn, 2002). The EV71 coding region contains three distinct areas, namely, P1, P2, and P3 (Chong et al., 2015). The P1 area produces four structural proteins, namely, VP1, VP2, VP3, and VP4. The P2 and P3 areas encode seven non-structural proteins, namely, 2A-2C and 3A-3D, the P2 and P3 areas encode seven non-structural proteins, namely, 2A-2C and 3A-3D (Wang et al., 2012; Solomon et al., 2010). EV71 is the major contributor to hand, foot, and mouth disease (HFMD), leading to seasonal morbidity and mortality in children aged 5 years and below. Its clinical characterized by lesions of the fever, rash, blisters and oral ulcers (Chan et al., 2000; Nolan et al., 2003). However, a minority of patients rapidly develop neurological disease, resulting in mortality and severe sequelae (Chang et al., 2007). HFMD depicted a strong seasonal pattern that mainly emerged in the spring (Deng et al., 2013), summer (Baek et al., 2011; Koh et al., 2016) and autumn (Van Tu et al., 2007; Lee et al., 2009). The incidence of HFMD is drastically higher in hot and humid conditions, relative to cold climate (Kim et al., 2016). This seasonal variation suggests that HFMD incidence might be influenced by environmental factors. The relationships between environmental factors and HFMD have been explored in some studies (Laor et al., 2020; Li et al., 2023; Tan et al., 2023).
EV71 was first isolated in 1969 from patients carrying central nervous system diseases (Ho et al., 1999). Subsequently, there were reports of frequent EV71 outbreaks in New York, Australia, Europe, and other countries (Dalldorf and Sickles, 1948). After 1997, EV71 produced the most casualties and widespread occurrences in the Asia-Pacific region (AbuBakar et al., 1999). In China, HFMD was first reported in Shanghai in 1981 (Xing et al., 2014), and EV71 was first isolated in Hubei in 1987. Over the past 20 years, significant progress has been made in the epidemiological and molecular biology research of HFMD in China. For instance, in the Fuyang City, Anhui Province reported 0.49 million cases of HFMD and 126 deaths in 2008 alone (Zhang et al., 2010). In the Chengdu region, a total of 29,861 laboratory-confirmed cases of enterovirus infections related to HFMD were reported from 2013 to 2022. This data highlights the prevalence and transmission patterns of HFMD in the area (Yang et al., 2023). In Hangzhou, Zhejiang Province, out of 560 samples collected between 2016 and 2022, 472 (84.29%) tested positive, indicating a high infection rate in the region. These findings not only underscore the intensity of HFMD spread in Hangzhou, but also emphasize the importance of continuous monitoring and prevention measures (Yuan et al., 2024). Furthermore, the Inner Mongolia Autonomous Region experienced an average annual incidence rate of HFMD of 71.99/100,000 from 2009 to 2018, which was lower than the national average of 134.59/100,000 during the same period. This disparity may be attributed to Inner Mongolia’s relatively low population density, which could influence the disease’s transmission dynamics. In our study, we infected Vero cells with the Inner Mongolia EV71 C33 strain and conducted RNA sequencing analysis to elucidate potential mechanisms related to the pathogenesis of EV71-associated HFMD. These research outcomes provide valuable insights into the molecular biology of HFMD and contribute to the improvement of disease management and prevention strategies. Through these comprehensive studies, we can gain a more comprehensive understanding of the epidemiological characteristics and potential pathogenic mechanisms of HFMD, offering scientific evidence to inform public health decision-making.
Materials and methods
Cell culture
The African green monkey kidney cells, known as Vero cells, were sourced from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, United States) supplemented with 10% fetal bovine serum (FBS, Gibco, United States), and 1% penicillin-streptomycin (Gibco, United States). This specific cell line is widely used in virology research due to its susceptibility to various viruses, including the EV71 C33λ strain, which was isolated from patients suffering from HFMD in Ordos (population of the district in Inner Mongolia). To initiate the experiment, Vero cells were seeded into a 6-well plate and introduced with 200 μL of the EV71 C33λ virus, achieving an infection titer of 106.0TCID50 per well. The cells were cultured in a wet chamber of 37°C with 5% CO₂. The cells were then incubated in a humidified chamber at 37°C with 5% CO₂ to facilitate viral replication and the development of cytopathic effects. Once the cells exhibited clear signs of necrosis and viral shedding, indicative of successful infection, total RNA was extracted from both the control and experimental groups.
Total RNA extraction
Total viral RNA isolation was done from cells using TRIzol™ Reagent Kit (Invitrogen, Carlsbad, CA, United States), as per kit directions. RNA quantification was done via NanoDrop2000 (Thermo Fisher Scientific, Waltham, MA, United States), eukaryotic mRNA was enriched by Oligo(dT) NA beads, while prokaryotic mRNA was enriched by removing rRNA by Ribo-Zero™ Magnetic Kit (Epicentre, Madison, WI, United States). Then the enriched mRNA was fragmented into Kit (Epicentre, Madison, WI, United States). Then the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse transcripted into cDNA with random primers. Second-strand cDNA were synthesized by DNA polymerase I, RNase H, dNTP and buffer. Then the cDNA fragments were purified with QiaQuick PCR extraction kit (Qiagen, Venlo, Netherlands), end repaired, poly (A) added, and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeq2500 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Quality filtering and map reading
To ensure data quality, it was necessary to filter the raw data before information analysis in order to reduce the interference caused by invalid data. First, we utilized fastp (Chen et al., 2018) to perform quality control on the raw reads obtained from sequencing, filtering out low-quality data (filtering criteria detailed below), resulting in clean reads.
Sequence data analysis
Reads with special mapping were utilized for further information. RNA-Seq of all samples was measured using differentially expressed genes (DESeq2) (Love et al., 2014). The same method was applied to estimate differences between contrasting genotypes. Based on our differential analysis of RNA-Seq data, genes were identified as significantly different if they exhibited a False Discovery Rate (FDR) < 0.05 and |log2FC| (fold change) > 1.
Functional classification and pathway identification
Principal component analysis (PCA) was conducted using R1 to study the distance relationships between samples through dimension reduction. Subsequently, functional enrichment analysis of differentially expressed genes (DEGs) was performed using GO2 and KEGG.3
Reverse transcription quantitative PCR (RT-qPCR) confirmation
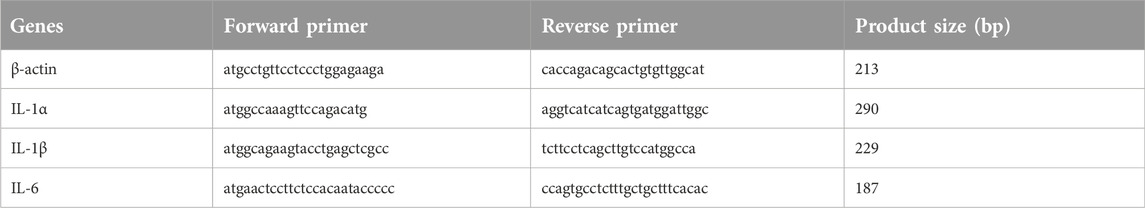
Three differential genes were identified by RT-qPCR. The following genes had elevated expression: interleukin 1-alpha (IL1α, GenBank: NM000575.5), interleukin 1-beta (IL1β, GenBank: NM000576.3), and interleukin-6 (IL-6, GenBank: NM001371096.1). The employed PCR primers were acquired from Sangon Biotech. The designed primers are summarized in Table 1. The RT-qPCR reaction was done as follows: cDNA 1 μL, forward and reverse primers (0.4 µL each), 2×SYBR Green Master Mix 10 μL, ddH2O 8.2 µL, and in total volume of 20 µL. The PCR amplifications were performed using a CFX96 Touch Real-Time PCR Detection System (C1000Touch™ Thermal Cycler, BIO-RAD). The amplification curve reaction program: 98°C for 2 min, 95°C for 10 min, 95°C for 15 s, 58°C for 1 min, 30 cycles; Melting curve at 95°C for 10 min, 60°C for 30 s, and 95°C for 10 min. Finally, all data were statistically analyzed using t-test (SPSS ver.19.0) and are depicted as mean ± SD. When p < 0.05, the effect was considered significant. We repeated the tests for three times each sample.
Results
Gene annotation
RNA-Seq was a rapid acquisition of all transcripts of a species-specific organ or cell in a certain state, using high-throughput sequencing technology. The result reflects the expression levels of all transcripts. In this study, we analyzed the transcriptome responses of Vero cells infected with the EV71 C33λ strain, using RNA-Seq. Overall, a total of 42,088,874 unigenes (non-infected control) were annotated against CleanDate (41,977,800 unigenes/99.74%) and LowQuality (109,116 unigenes/0.26%) using BLASTx, respectively (E-value < 1 × 10−5). A total of 42,357,062 unigenes (EV71 C33λ group) were annotated against CleanDate (42,258,546 unigenes/99.77%) and LowQuality (96,960 unigenes/0.23%) using BLASTx, respectively. The average cleanliness rate of reads was >90%, indicating good reliability of the RNA-Seq data.
Gene expression pattern analysis
Vero cells were infected with EV71 C33λ (106.0TCID50), prior to total RNA extraction and RNA-Seq (Omicsmart platform). Principle Component Analysis (PCA) was performed on all gene counts (Figure 1A). The value of PCA was PC1 = 63.7%, PC2 = 24.8%. Based on the RNA-Seq analysis, we identified genes with FDR < 0.05 and |log2FC| > 1 as DEGs. We identified 942 DEGs, among which, 568 had high expression, and 374 had low expression (Figure 1B).
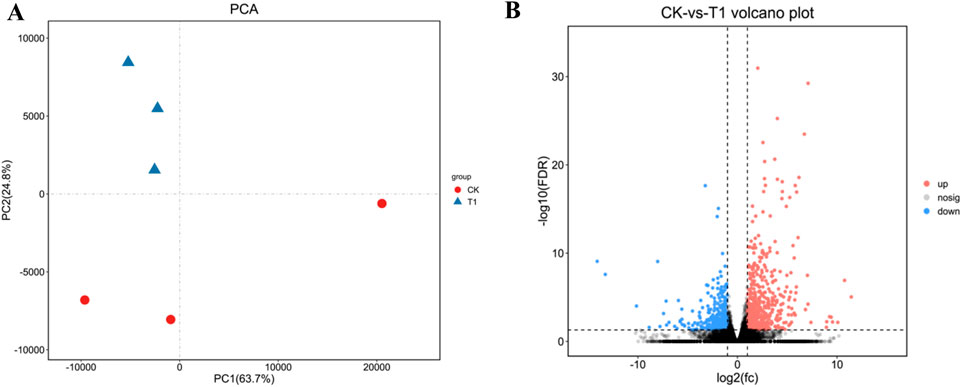
Figure 1. The visualization of the sample relationship for differently expressed genes (DEGs). (A) Principal Component Analysis (PCA); where samples are distinguished by different colors based on their effective treatments. (B) A Volcano map; which uses red to indicate genes with elevated levels and blue to indicate those with reduced expression levels.
GO analysis of DEGs
To elucidate the activity of differentially expressed genes (DEGs), we conducted a gene ontology (GO) enrichment analysis on these genes before and after viral infections. GO encompasses three main categories: “biological process,” “cellular component,” and “molecular function” (Figure 2). Within the “biological process” category, 3,541 genes were identified with high expression levels, while 1,702 genes exhibited low expression. Regarding the “cellular component” 1,839 genes were found to be highly expressed, and 1,074 genes were expressed at lower levels. The most enriched terms in this category include “cell” (540 genes, 18.53%), “cell part” (539 genes, 18.50%), “organelle” (375 genes, 12.87%), and “membrane” (351 genes, 12.04%). In the “molecular function” category, 705 genes were highly expressed, and 425 genes were expressed at lower levels, with “binding” (569 genes) being the most prevalent term at 50.35%, followed by “catalytic activity” (252 genes) at 22.30%.
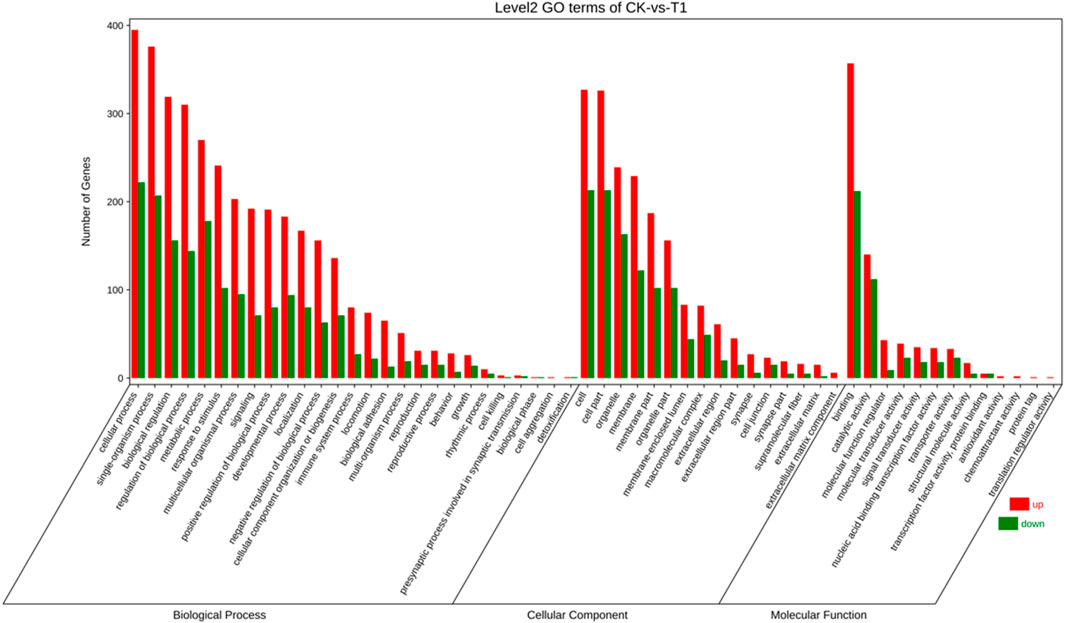
Figure 2. The gene ontology (GO) enrichment classification of EV71 C33λ-mediated viral infection encompasses biological processes (BP), cellular components (CC), and molecular function (MF).
The abscissa denotes the second-level GO terms, while the ordinate indicates the count of genes within each term. Elevated levels of gene expression are indicated by red, and reduced levels are indicated by green.
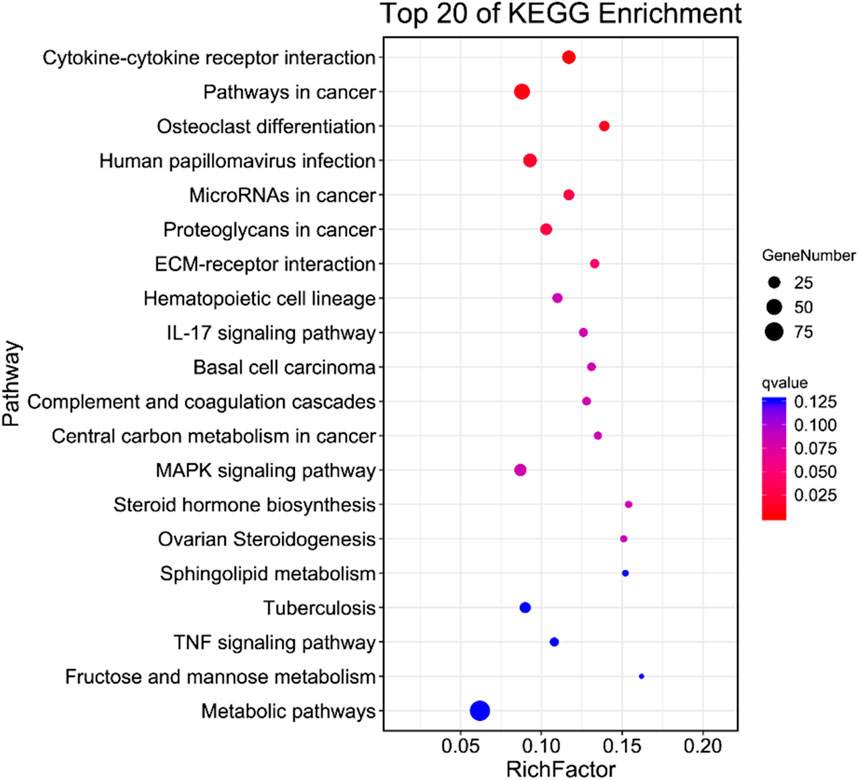
KEGG pathway enrichment analysis
To delve into the physiological functions of DEGs, we conducted an enrichment analysis on the KEGG pathway annotation of on the DEGs from the virus Control (CK) vs. Treatment (T1). Our analysis revealed that the enrichments of DEGs was primarily associated with human diseases, metabolism, biological systems, genetic information transmission, cellular processes, and physiological processes, including environmental information processing (Figure 3). Additionally, a significant enrichment was observed in cytokines and cytokine receptors. This suggests that upon EV71 infection in Vero cells, there was a substantial alteration in the expression of genes related to cytokines and cytokine receptors which in turn elicited a robust immune response.
RT-qPCR analysis
The analysis of the KEGG classification has identified the primary upregulated genes associated with human diseases, which will be further investigated. In the context of EV71-inoculated Vero cells, a significant upregulation in the expression levels of immune molecules, specifically IL-1α, IL-1β, and IL-6, was observed when compared to the control group. This robust increase in immune molecule expression suggests that the cellular immune response was activated in response to the EV71 virus infection (Figure 4).
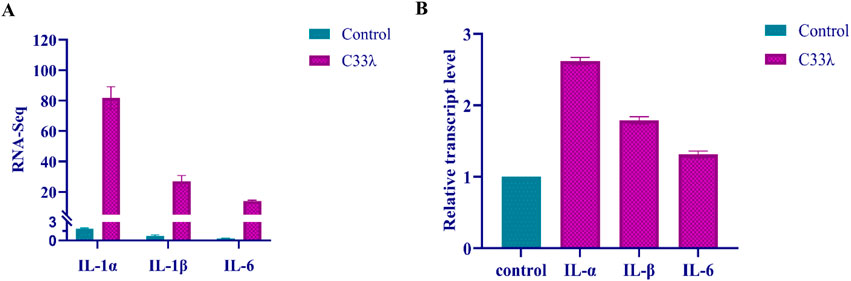
Figure 4. Illustrates the quantification of IL-1α, IL-1β, and IL-6 expression in Inner Mongolia EV71 C33λ strain from EV71-inoculated Vero cells, as determined by both RNA-Seq (RNA sequencing) and RT-qPCR (quantitative real-time PCR) techniques. (A) Displays the quantification of IL-1α, IL-1β, and IL-6 in Inner Mongolia EV71 C33λ strain using RNA-Seq; (B) shows the quantification of these cytokines via RT-qPCR. Each RT-qPCR reaction was replicated three times for each sample, and the standard errors are represented by the vertical bars.
Discussion
Enterovirus 71 (EV71) is a significant pathogen that causes HFMD in children aged 5 years or younger (Lee, 2016). Consequently, its research is of substantial societal importance. Despite the EV71 vaccine commendable safety profile and high efficacy, it does not provide therapeutic benefit to patients who have already contracted EV71. Therefore, unraveling the pathogenesis of EV71 is particularly critical for the wellbeing of infants and young children. Here, we scrutinized the RNA-seq data from EV71-infected Vero cells to elucidate the pathogenic mechanisms of EV71 with compelling evidence. Our investigation has uncovered a significant alteration in gene expression profiles within EV71-infected Vero cell, with 568 genes exhibiting increased expression and 374 genes showing decreased expression. We hypothesize that these differentially expressed genes (DEGs) play a pivotal role in the progression of EV71 infection. To elucidate the potential biological roles of these DEGs, we performed comprehensive KEGG and GO functional enrichment analyses. Focusing on the “cytokine and cytokine receptor” pathway, our pathway analysis highlighted the enrichment of IL-1α, IL-1β, and IL-6 within this critical signaling network, suggesting their potential as transcriptional regulators. During EV71 infection, these genes were found to be robustly upregulated, indicating their importance in the host’s immune response to the virus.
IL-1α, pivotal cytokine in immune regulation, requires the enzymatic activity of calpain to exert its full function. This cytokine is synthesized in response to the activation of inflammasomes and during the process of cellular senescence. Shi et al. (2014) has demonstrated that EV71 infection leads to a significant elevation in IL-1α expression, with levels ranging from 2.04 to 5.49 times higher than those observed in uninfected controls. Furthermore, the application of luminex fluorescence technology has confirmed that EV71 infection induces a marked increase in IL-1α secretion compared to non-infected cells. These findings underscore the critical role of IL-1α in the body’s immune response to EV71 infection and highlight the potential of targeting IL-1α pathways for the development of therapeutic strategies aimed at managing related inflammatory conditions. Our findings revealed that EV71 infection led to a significant elevation in IL-1α expression, with levels approximately doubling compared to the control group. IL-1β, a pro-inflammatory cytokine, primarily targets monocytes, endothelial cells, and fibroblasts to initiate an immune response (van Bon et al., 2011). It is pivotal in both systemic and local reactions to infections, agents, physical injuries, and immune challenges, and is a central player in the development of both chronic and acute inflammatory conditions (Dinarello, 1998). Griffiths et al. (2012) observed that patients with EV71-infection exhibit increased IL-1β levels compared to those in healthy patients. Our research indicates that IL-1β expression progressively rises following EV71 infection. These observations suggest that IL-1β may be instrumental in the pathogenesis of EV71-induced HFMD and its associated complications. Similarly, IL-6, another pro-inflammatory cytokine, is typically expressed at low levels during development but is markedly upregulated in inflammatory states. Research has shown that EV71 infection leads to a significant increase in IL-6 expression and secretion (Huang et al., 2011). Treatment with anti-IL-6 antibodies in mice has been associated with reduced tissue damage (Khong et al., 2011). Our findings corroborate these results, demonstrating a substantial upregulation of IL-6 levels in Vero cells EV71 infection. Consequently, we propose that IL-6 also plays a critical role in the progression of EV71 infection.
In conclusion, we have documented the RNA-Seq profiles of Vero cells infected with the EV71 C33λ strain. Our data will serve as a cornerstone for future comprehensive studies into the mechanisms by which inflammatory factor contribute to the pathogenesis of EV71 C33λ infection.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal study was approved by Department of Science and Technology of Inner Mongolia Autonomous Region. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RF, BL, and WX is the principal investigator of this study, XT, XaL, YQ, and SG conceived and designed the study. XZ, HuL, XY, HoL, JM, XJ, WZ, DW, and YS performed all experiments. XoL and YF wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Inner Mongolia Natural Science Foundation (Grant Numbers: 2024MS03039; 2019MS03027); Inner Mongolia Health Science and Technology Project (Grant Number: 202201156); The “Flower Bud Project” project of Baotou Medical College (Grant Number: HLJH202419).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
AbuBakar, S., Chee, H. Y., Al-Kobaisi, M. F., Xiaoshan, J., Bing Chua, K., and Kit Lam, S. (1999). Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 61 (1), 1–9. doi:10.1016/s0168-1702(99)00019-2
Baek, K., Yeo, S., Lee, B., Park, K., Song, J., Yu, J., et al. (2011). Epidemics of enterovirus infection in Chungnam Korea, 2008 and 2009. Virology J. 8, 297. doi:10.1186/1743-422x-8-297
Chan, L. G., Parashar, U. D., Lye, M. S., Ong, F. G. L., Zaki, S. R., Alexander, J. P., et al. (2000). Deaths of children during an outbreak of hand, foot, and mouth disease in Sarawak, Malaysia: clinical and pathological characteristics of the disease. Clin. Infect. Dis. 31 (3), 678–683. doi:10.1086/314032
Chang, L. Y., Huang, L. M., Gau, S. S., Wu, Y. Y., Hsia, S. H., Fan, T. Y., et al. (2007). Neurodevelopment and cognition in children after enterovirus 71 infection. N. Engl. J. Med. 356 (12), 1226–1234. doi:10.1056/NEJMoa065954
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinforma. Oxf. Engl. 34 (17), 884–890. doi:10.1093/bioinformatics/bty560
Chong, P., Liu, C. C., Chow, Y. H., Chou, A. H., and Klein, M. (2015). Review of enterovirus 71 vaccines. Clin. Infect. Dis. 60 (5), 797–803. doi:10.1093/cid/ciu852
Dalldorf, G., and Sickles, G. M. (1948). An unidentified, filtrable agent isolated from the feces of children with paralysis. Sci. (New York, NY) 108 (2794), 61–62. doi:10.1126/science.108.2794.61
Deng, T., Huang, Y., Yu, S., Gu, J., Huang, C., Xiao, G., et al. (2013). Spatial-temporal clusters and risk factors of hand, foot, and mouth disease at the district level in Guangdong Province, China. PLoS One 8 (2), e56943. doi:10.1371/journal.pone.0056943
Dinarello, C. A. (1998). Interleukin-1β, interleukin-18, and the interleukin-1β converting enzymea. Ann. N. Y. Acad. Sci. 856, 1–11. doi:10.1111/j.1749-6632.1998.tb08307.x
Griffiths, M. J., Ooi, M. H., Wong, S. C., Mohan, A., Podin, Y., Perera, D., et al. (2012). In enterovirus 71 encephalitis with cardio-respiratory compromise, elevated interleukin 1, interleukin 1 receptor antagonist, and granulocyte colony-stimulating factor levels are markers of poor prognosis. J. Infect. Dis. 206 (6), 881–892. doi:10.1093/infdis/jis446
Ho, M., Chen, E. R., Hsu, K. H., Twu, S. J., Chen, K. T., Tsai, S. F., et al. (1999). An epidemic of enterovirus 71 infection in taiwan. N. Engl. J. Med. 341 (13), 929–935. doi:10.1056/nejm199909233411301
Huang, S. W., Lee, Y. P., Hung, Y. T., Lin, C. H., Chuang, J. I., Lei, H. Y., et al. (2011). Exogenous interleukin-6, interleukin-13, and interferon-γ provoke pulmonary abnormality with mild edema in enterovirus 71-infected mice. Respir. Res. 12 (1), 147–152. doi:10.1186/1465-9921-12-147
Khong, W. X., Foo, D. G., Trasti, S. L., Tan, E. L., and Alonso, S. (2011). Sustained high levels of interleukin-6 contribute to the pathogenesis of enterovirus 71 in a neonate mouse model. J. Virology 85 (7), 3067–3076. doi:10.1128/jvi.01779-10
Kim, B. I., Ki, H., Park, S., Cho, E., and Chun, B. C. (2016). Effect of climatic factors on hand, foot, and mouth disease in South Korea, 2010-2013. PLoS One 11 (6), e0157500. doi:10.1371/journal.pone.0157500
Koh, W. M., Bogich, T., Siegel, K., Jin, J., Chong, E. Y., Tan, C. Y., et al. (2016). The epidemiology of hand, foot and mouth disease in Asia: a systematic review and analysis. Pediatr. Infect. Dis. J. 35 (10), e285–e300. doi:10.1097/inf.0000000000001242
Laor, P., Apidechkul, T., Khunthason, S., Keawdounglek, V., Sudsandee, S., Fakkaew, K., et al. (2020). Association of environmental factors and high HFMD occurrence in Northern Thailand. BMC Public Health 20 (1), 1829. doi:10.1186/s12889-020-09905-w
Lee, K. Y. (2016). Enterovirus 71 infection and neurological complications. Korean J. Pediatr. 59 (10), 395–401. doi:10.3345/kjp.2016.59.10.395
Lee, T. C., Guo, H. R., Su, H. J., Yang, Y. C., Chang, H. L., and Chen, K. T. (2009). Diseases caused by enterovirus 71 infection. Pediatr. Infect. Dis. J. 28 (10), 904–910. doi:10.1097/INF.0b013e3181a41d63
Li, C. H., Mao, J. J., Wu, Y. J., Zhang, B., Zhuang, X., Qin, G., et al. (2023). Combined impacts of environmental and socioeconomic covariates on HFMD risk in China: a spatiotemporal heterogeneous perspective. PLoS neglected Trop. Dis. 17 (5), e0011286. doi:10.1371/journal.pntd.0011286
Liu, C. C., Chou, A. H., Lien, S. P., Lin, H. Y., Liu, S. J., Chang, J. Y., et al. (2011). Identification and characterization of a cross-neutralization epitope of Enterovirus 71. Vaccine 29 (26), 4362–4372. doi:10.1016/j.vaccine.2011.04.010
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550–571. doi:10.1186/s13059-014-0550-8
McMinn, P. C. (2002). An overview of the evolution of enterovirus 71 and its clinical and public health significance. Fems Microbiol. Rev. 26 (1), 91–107. doi:10.1111/j.1574-6976.2002.tb00601.x
Nolan, M. A., Craig, M. E., Lahra, M. M., Rawlinson, W. D., Prager, P. C., Williams, G. D., et al. (2003). Survival after pulmonary edema due to enterovirus 71 encephalitis. Neurology 60 (10), 1651–1656. doi:10.1212/01.wnl.0000066810.62490.ff
Oprisan, G., Combiescu, M., Guillot, S., Caro, V., Combiescu, A., Delpeyroux, F., et al. (2002). Natural genetic recombination between co-circulating heterotypic enteroviruses. J. general virology 83 (9), 2193–2200. doi:10.1099/0022-1317-83-9-2193
Shi, W., Hou, X., Peng, H., Zhang, L., Li, Y., Gu, Z., et al. (2014). MEK/ERK signaling pathway is required for enterovirus 71 replication in immature dendritic cells. Virology J. 11, 227–240. doi:10.1186/s12985-014-0227-7
Solomon, T., Lewthwaite, P., Perera, D., Cardosa, M. J., McMinn, P., and Ooi, M. H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis., 2010, 10(11): 778–790. doi:10.1016/s1473-3099(10)70194-8
Tan, C., Li, S., Li, Y., and Peng, Z. (2023). Dynamic modeling and data fitting of climatic and environmental factors and people's behavior factors on hand, foot, and mouth disease (HFMD) in Shanghai, China. Heliyon 9 (8), e18212. doi:10.1016/j.heliyon.2023.e18212
van Bon, L., Cossu, M., and Radstake, T. R. (2011). An update on an immune system that goes awry in systemic sclerosis. Curr. Opin. rheumatology 23 (6), 505–510. doi:10.1097/BOR.0b013e32834b0dac
Van Tu, P., Thao, N. T. T., Perera, D., Truong, K. H., Tien, N. T. K., Thuong, T. C., et al. (2007). Epidemiologic and virologic investigation of hand, foot, and mouth disease, southern Vietnam, 2005. Emerg. Infect. Dis. 13 (11), 1733–1741. doi:10.3201/eid1311.070632
Wang, X., Peng, W., Ren, J., Hu, Z., Xu, J., Lou, Z., et al. (2012). A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. and Mol. Biol. 19 (4), 424–429. doi:10.1038/nsmb.2255
Xing, W., Liao, Q., Viboud, C., Zhang, J., Sun, J., Wu, J. T., et al. (2014). Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect. Dis. 14 (4), 308–318. doi:10.1016/s1473-3099(13)70342-6
Yang, Q., Liu, F., Chang, L., Lai, S., Teng, J., Duan, J., et al. (2023). Molecular epidemiology and clinical characteristics of enteroviruses associated HFMD in Chengdu, China, 2013-2022. Virology J. 20 (1), 202. doi:10.1186/s12985-023-02169-x
Yuan, Y., Chen, Y., Huang, J., Bao, X., Shen, W., Sun, Y., et al. (2024). Epidemiological and etiological investigations of hand, foot, and mouth disease in Jiashan, northeastern Zhejiang Province, China, during 2016 to 2022. Front. Public Health 12, 1377861. doi:10.3389/fpubh.2024.1377861
Keywords: enterovirus 71, HFMD, Verda Reno African green monkey kidney cell, RNA-seq, DEGs
Citation: Lai X, Fan Y, Li H, Tian X, Yue X, Gao S, Lei X, Qin Y, Zhang X, Mei J, Shi Y, Li H, Wu D, Zhang W, Jia X, Fan R, Li B and Xing W (2024) A RNA-seq-based study on differentially expressed genes related to the Inner Mongolia EV71 C33λ strain that invades Vero cells. Acta Virol. 68:12084. doi: 10.3389/av.2024.12084
Received: 21 September 2023; Accepted: 15 August 2024;
Published: 20 November 2024.
Edited by:
Ludovit Skultety, Institute of Virology (SAS), SlovakiaReviewed by:
Petr Pajer, Institute of Molecular Genetics (ASCR), CzechiaDiana Hopková, Slovak Academy of Sciences, Slovakia
Copyright © 2024 Lai, Fan, Li, Tian, Yue, Gao, Lei, Qin, Zhang, Mei, Shi, Li, Wu, Zhang, Jia, Fan, Li and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruichun Fan, MjQ3Mjc1MjIwODlAcXEuY29t; Bin Li, MzQzMzAwMTY2MkBxcS5jb20=; Wanjin Xing, eHdhbmppbkBpbXUuZWR1LmNu
†These authors have contributed equally to this work
 Xiong Lai1,2†
Xiong Lai1,2† Xuanzhi Yue
Xuanzhi Yue