- 1Enterovirus Laboratory, Institute of Microbiology, Faculty of Medicine, Slovak Medical University, Bratislava, Slovakia
- 2Department of Microbiology and Virology, Faculty of Natural sciences, Comenius University, Bratislava, Slovakia
- 3Viral Hepatitis Laboratory, Institute of Microbiology, Faculty of Medicine, Slovak Medical University, Bratislava, Slovakia
- 4Department of Medical Microbiology, Regional Authority of Public Health, Banska Bystrica, Slovakia
- 5National Reference Center for Polio, Department of Medical Microbiology, Public Health Office of the Slovak Republic, Bratislava, Slovakia
- 6Department of Biomedicine, University of Turku, Turku, Finland
- 7Department of Medical Microbiology, Radboud University Medical Centre, Nijmegen, Netherlands
Enterovirus (EV) infections occur frequently in humans. In some geographical areas they are more common. These viruses cause diseases with varying degrees of severity, from a simple respiratory tract infection to severe diseases. Since EVs include more than 70 serotypes currently circulating in the population, a methodology that detects most of them is needed. ELISA is a rapid, sensitive, and economical diagnostic method for the identification of EV serotypes and can also be used as a retrospective diagnostic tool or in the investigation of outbreaks of infection. Commercial EV-ELISAs often appear and gradually disappear from the market supply. We have used the KTL-510 peptide, a synthetic viral protein of poliovirus VP1, as an antigen in a peptide-based ELISA for the detection of a broader spectrum of anti-EV antibodies. We aimed to design, optimize, and standardize this in-house ELISA with the peptide, and implement the method for routine detection of anti-EV IgG in human sera. For determining the cut-off value, we used 100 patients’ sera which were previously tested negative for IgG antibodies against EVs using a commercial ELISA kit available. We monitored patients’ sera samples sent for serological testing of anti-coxsackievirus antibodies to the National Reference Center for the Identification of Enteric Viruses between 2018–2022. These serum samples were examined using a standard virus neutralization test as well as the newly developed ELISA method.
Introduction
Genus Enterovirus which belongs to the Picornaviridae family includes numerous viral species widespread in the human population. They are most often disseminated by the fecal-oral route, less often by the respiratory route, and infection rates vary according to the location and season. Enteroviruses (EVs) occur commonly, though often the infections go unnoticed. They are the etiological agents of hand, foot, and mouth disease, respiratory tract infections, herpangina, and pleurodynia. In some cases, they can lead to life-threatening diseases of the central nervous system, severe infections with permanent consequences, or death. They may play a role in some chronic diseases such as chronic myocarditis, dilated cardiomyopathy, post-polio syndrome, or type 1 diabetes. Children, the elderly, and people with weakened immunity are the most endangered by EV infection (Chapman and Kim, 2008; Pallansch et al., 2013; Smatti et al., 2019; Itani et al., 2023). Human EVs are divided into four EV species (A to D) and three rhinovirus species (A to C) based on sequence diversity. Currently, 75 EV serotypes and 100 rhinovirus serotypes are known according to their antigenicity, and more than 300 types have been characterized based on their genetics (International Committee on Taxonomy of Viruses, 2023).
Therefore, it is difficult to find a suitable method for their diagnosis that would be fast, specific, and sensitive, while identifying a large number of EV species. Methods, such as virus isolation, can only reveal acute EV infection. In contrast, serological methods can be used to diagnose both acute and chronic EV infections. Serological tests used in the routine diagnosis of EVs include complement fixation test, enzyme-linked immunosorbent assay (ELISA), or virus neutralization test (VNT) (Swanink et al., 1993; Payne, 2017), the latter being used in our laboratory. The biggest disadvantage of VNT is the time consumed to get the results. It captures antibodies against specific (selected) serotypes, which may be of interest for epidemiological studies, but when detecting many serotypes, this method is financially and time inefficient.
Advantages of the ELISA is that it is rapid, sensitive, it can be automated and is useful for the detection of acute and chronic infections, in retrospective diagnosis and outbreak studies. Several EV serotypes can be detected in ELISAs capturing heterotypic antibodies using antigens from a limited number of EVs or synthetic peptide antigens in a single run (Craig et al., 2003; Payne, 2017). M. Roivainen and her group identified regions in the poliovirus (PV) type 3 capsid proteins that bind anti-EV antibodies using a peptide scanning method. Such antibody binding sites were found in regions of the capsid virus protein (VP) 1, VP2, and VP3. VP1 protein (located between amino acids 37 and 53 of the polypeptide chain) was identified with the highest degree of sequence homology in all EVs analyzed (PV1 Mahoney and Sabin, PV2 Lansing and Sabin, PV3 Sabin, Finland/84 and Leon, coxsackievirus (CV) A9, A21, B1, B3, B4, and B5, EV71) in the study except echovirus (ECV) 22 (Roivainen et al., 1991), which has been reclassified as a member of the Parechovirus genus (International Committee on Taxonomy of Viruses, 2023). Based on this region, 4 peptides were synthesized. Rabbits were experimentally immunized using all these synthetic peptides, and the sera were further analyzed. Authors Hovi and Roivainen tested different members of the genus Enterovirus used immunoperoxidase in situ hybridization assay. The most effective peptide was KTL-510 with the sequence KEVPALTAVETGAT(C), which covered several EV serotypes (PV type 1–3, CVA9, A13, A16, A18, A21, CVB1 - B6, 23 different serotypes of ECV and EV69, EV70, and EV71). The peptide did not react with CVA7 (EV) and also with other testes DNA and RNA viruses. The study did not examine EVs other than those previously mentioned (Hovi and Roivainen, 1993). We assume that the changes in the amino acid sequence of this target region of the VP1 capsid protein in some specific serotypes (e.g., mutations) may act as a limiting factor in the range of detection.
Based on the broad EV detection spectrum and specificity to VP1 region, we used the KTL-510 peptide as an antigen in a peptide-based ELISA to detect EV immunoglobulin (Ig) G in our study. Our aim was to design, optimize, and standardize a peptide-based ELISA and implement it into the routine diagnosis of IgG against EV in human sera.
Material a methods
Serum samples
The Office of Public Health (Bratislava, SR) kindly provided us with 100 human sera samples that were evaluated in their laboratory as anti-EV IgG negative by a commercial ELISA kit. Another set of 109 samples was evaluated with VNT as negative for selected anti-CV antibodies in the Regional Office of Public Health (Banská Bystrica, SR) and the National Reference Center for the Identification of Enteric Viruses (Bratislava, SR). These tested sera were used to establish a cut-off value, which was determined based on the optical density (OD) values of at least 100 negative sera diluted 1:1,000.
For ELISA and VNT testing, we used 402 human sera sent to the National Reference Center for the Identification of Enteric Viruses (Bratislava, SR) for CV antibody examination.
Enzyme-linked immunosorbent assay for detection of anti-EV IgG antibodies
Round-bottom 96-well plates were coated with KTL-510 peptide at a concentration of 5 μg/mL (Roivainen et al., 1998) in 0.05 M carbonate buffer (pH 9.0) and incubated overnight (maximum of 5 days) at 8°C. After binding, the wells were washed three times with phosphate buffered saline (PBS) enriched by 0.1% Tween 20, the washing was repeated after almost every subsequent step. Coated plates were blocked with PBS with 0.1% bovine serum albumin for 30 min at room temperature. Although most commercial ELISA kits do not require serum inactivation, it is a standard step for many serological methods. We assessed (compared) inactivated and non-inactivated sera. The sera were heat-inactivated at 56°C or 50°C for 30 min in a water bath, then diluted in PFTM buffer (PBS with 1% fetal bovine serum, 1% bovine serum albumin, 0.1% Tween 20, and 5 mM ethylenediaminetetraacetic acid) in a ratio of 1:100, 1:1,000 and 1:10000. Human sera were analyzed in duplicates. To peptide-bound plates, different dilutions of serum samples were added, incubated for 1 h at 36°C and then washed three times. Polyclonal rabbit antibody against human IgG labelled with horse radish peroxidase (DAKO, P02104) diluted in PFTM buffer (1:6,000) was applied to the plates and incubated for 1 h at 36°C, then washed (3x). Finally, the substrate containing urea-peroxide, sodium citrate, sodium acetate trihydrate, and 3,3′,5,5′-Tetramethylbenzidine, pH 5.5) was added. After 10-min incubation time, the reaction was stopped with 1 N sulfuric acid. The OD values of individual wells were determined using iMark™ Microplate Reader (Bio-Rad Laboratories, Inc.) at a 450 nm wavelength (Supplementary Material).
Virus neutralization test for detection of anti-CV neutralizing antibodies
Neutralizing antibodies against CVB1, CVB2, CVB3, CVB4, CVB5, CVB6, CVA7, and CVA9 in two paired sera samples from one patient taken at a time interval of 2-3 weeks were measured using VNT. Serial 10-fold dilutions of virus strains (ranging from 10−1–10−8) in Eagles’ Minimum Essential Medium were prepared. Patient sera were diluted as two-fold dilutions (1:8; 1:16; 1:32; 1:64; 1:128, and 1:256) in 96-well plates. An equal volume (25 µL) of diluted CV serotype suspension (one selected serotype per row of microtiter plate) was added to diluted serum samples (inactivated at 56°C for 30 min). The dilution of individual viruses used in the experiment was 2-3 log10 of the viral titer to ensure that the virus reliably causes a cytopathic effect on the used cell line. After virus-serum incubation (1 h at 37°C), 50 µL/well of Vero cells (106 cells/plate) in Eagles’ Minimum Essential Medium containing 5% FBS was added, and after 5 days in a 5% CO2 atmosphere at 37°C evaluated. Uninfected cells (negative controls), controls of serum toxicity, and individual virus titration were included. The cytopathic effect (CPE) was observed and recorded, and the titer of neutralizing antibodies against single CV serotypes (barrier to CPE formation) was determined.
Results
Heat-inactivation
To determine whether inactivation was necessary, each non-inactivated sample was divided into 3 parts: one part remained non-inactivated, and the other parts were inactivated at 56°C or at 50°C for 30 min in a water bath. However, a relevant comparison of the sample processing methods could not be performed, as a large part of the samples (tested VNTs) were already inactivated at 56°C for 30 min (before starting experimental work), or we did not have sufficient volume to process the sample by all three methods. For data comparison, we had a relatively small set (n = 62) of non-inactivated and inactivated samples. Based on these results (not published data) and recommendations from the available literature, inactivation at 56°C for 30 min was chosen as the standard procedure. Using this inactivation protocol, we processed all serum samples tested, including those used to establish the cut-off. During this analysis, we found that thermal inactivation of human serum affected the measured absorbance, while the OD value increased with increasing temperature.
Cut-off determination
According to the 68-95-99.7 rule, observed data for a normal distribution will fall within 3 standard deviations (SD) of the average. Statistical rule states that 99.7% of the data should fall within 3 SD, 95% of the data within 2 SD and 68% of the data within 1 SD of the mean (Hubert, 2018). In the available literature, the cut-off value is calculated as the average value of negative samples OD plus 2 SD (mean + 2 SD) or mean plus 3 SD (mean + 3 SD) (Cao et al., 2022; Hochman et al., 2023; Oluka et al., 2023).
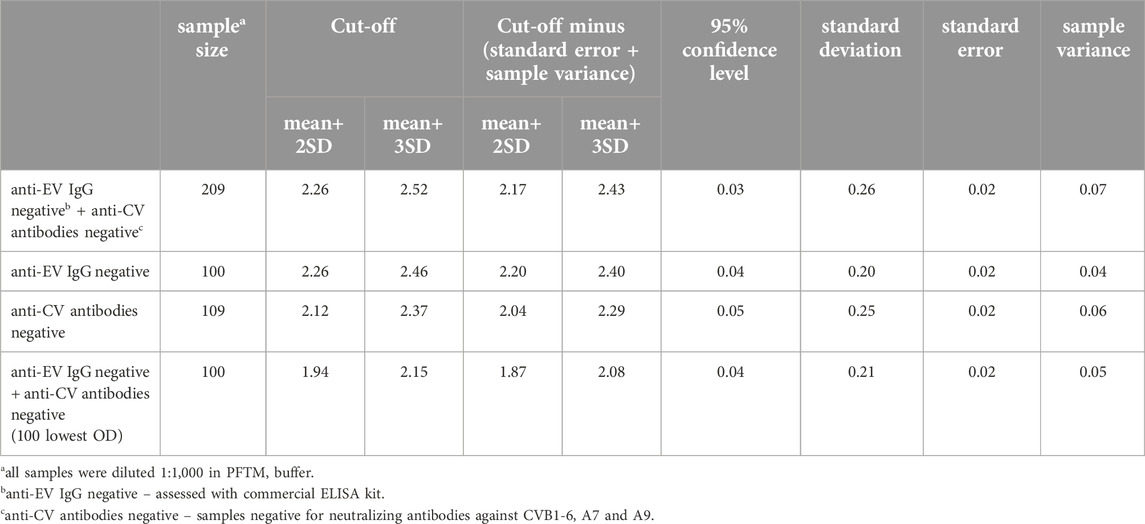
We used four sets of samples (diluted 1:1,000) to determine the cut-off value (mean + 2 SD and mean + 3 SD), 95% confidence level, standard deviation, standard error, and sample variance, as seen in Table 1. We examined 209 serum samples (inactivated before at 56°C for 30 min), of these 100 serum samples using the anti-EV IgG negative by commercial ELISA kit and 109 serum samples evaluated with the VNT for were anti-CV B1-6, A7 and A9 negative. We evaluated the cut-off value for the entire set of samples (n = 209), where the mean + 2 SD was 2.26 ± 0.03 and the mean + 3 SD was 2.52 ± 0.03. In the 100 serum samples (anti-EV IgG negative by commercial ELISA kit) we determined cut-off, the mean + 2 SD was 2.26 ± 0.04 and the mean + 3 SD was 2.46 ± 0.04. In the 109 serum samples (VNT tested as anti-CV B1-6, A7 and A9 negative) we determined cut-off, the mean + 2 SD was 2.12 ± 0.05 and the mean + 3 SD was 2.37 ± 0.05. Using VNT, only antibodies against selected CV strains were monitored, which is a limiting factor in determining the cut-off based on these samples. We used 100 serum samples with the lowest OD values from the entire sample set, for the determination of the cut-off value of the in-house ELISA. The mean + 2 SD were 1.94 ± 0.04 and mean + 3 SD 2.15 ± 0.04. Then the cut-off value of the in-house ELISA was determined as the average of the mentioned values i.e., 2.04 ± 0.04 (samples above 2.15 ± 0.04 were considered as highly positive, and samples between the cut-off and 1.94 ± 0.04 were in the upper limit of negativity). After subtracting the standard error and sampling variance from the set cut-offs, only minor differences were found in the numbers. The limitation of the optimization process is that the cut-off value was determined using 100 serum samples, which does not represent a diverse population and the range of anti-EV antibody levels.
Reproducibility within the laboratory work was determined using a single sample of human serum tested 35 times on different days and plates with time, laboratory temperature, and multiple freezing and thawing of the sample as varying factors. The average OD of the sample value was 2.09 with a SD of 0.15% and 95% confidence level of 0.05. According to these results, the coefficient of variation was 7.06%, which is acceptable because the satisfactory inter-assay precision is usually less than 10% (Murray and Lawrence, 1993).
Detection of anti-EV IgG in patients’ sera with in-house ELISA
We examined 402 patients’ sera samples sent for serological testing of anti-CV (B1-6, A7 and A9) antibodies to the National Reference Center in 2018–2021 using VNT and the newly developed ELISA method. These samples were not tested with commercial ELISA kit. This study was double blind. We evaluated the absorbance values from 1:1,000 dilutions and to assess positivity or negativity in ELISA, we used a set cut-off value. ELISA results are presented separately for each year in which the samples were taken. In 2021, 17/80 (21.3%) samples were positive (≥2.04 ± 0.04). Among the sera collected in 2020, 17/104 (16.3%) were positive. In 2019, 16/104 (15.4%) samples were positive. Out of the 114 sera from 2018, only 4 (3.5%) were positive. The OD values of several negative serum samples were in the upper limit of negativity (between ≥1.94 ± 0.04 and <2.04 ± 0.04), specifically 12/63 (19%) in 2021, 8/87 (9.2%) in 2020, 20/88 (22.7%) in 2019 and 6/110 (5.5%) of serum samples in 2018 (Figures 1, 2).
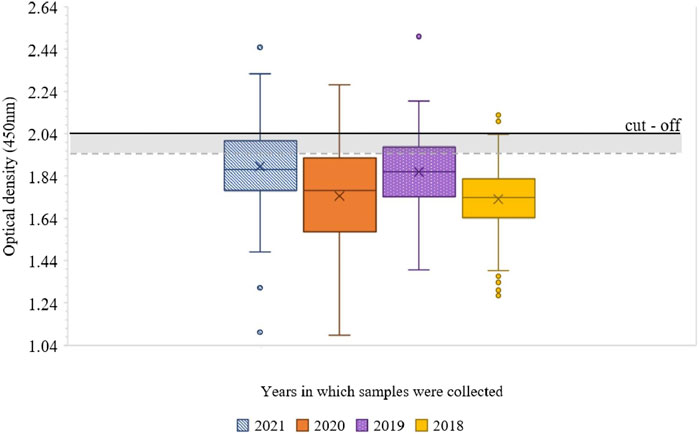
Figure 1. Boxplot of the distribution of examined human sera samples absorbance values at 450 nm, for each year in which the samples were taken. The cut-off value of 2.04 is shown in the graph as a linear black line and the lower limit of negativity as a grey dashed line. Negative samples are below <1.94 ± 0.04, the upper limit of negativity is between ≥1.94 ± 0.04 and <2.04 ± 0.04, and samples above the cut-off limit ≥2.04 ± 0.04 are positive.
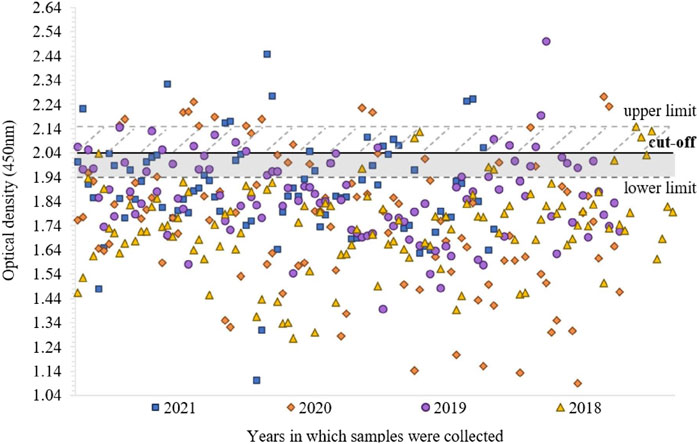
Figure 2. Distribution of absorbance values (at 450 nm) of 402 examined human sera samples, for each year in which the samples were taken. Negative samples are below <1.94 ± 0.04, the upper limit of negativity is between ≥1.94 ± 0.04 and <2.04 ± 0.04, samples above ≥2.04 ± 0.04 are positive and above ≥2.15 ± 0.04 highly positive.
Examination of patients’ sera with VNT
Based on VNT results, 152 out of 402 screened human sera were positive (antibody titer ≥128) for the presence of neutralizing antibodies against CV, namely anti-CVB1 in 12/152 (7.9%), anti-CVB2 in 11/152 (7.2%), anti-CVB3 in 50/152 (32.9%), anti-CVB4 in 64/152 (42.1%), anti-CVB5 in 50/152 (32.9%), anti-CVB6 only in 2/152 (1.3%), anti-CVA7 in 29/152 (19%) and anti-CVA9 in 48/152 (31.6%) sera. In 76 samples the antibody titer was ≥256. Some samples showed antibodies to several CVs. In Figure 3 we present the percentage of anti-CV neutralizing antibodies confirmed samples from the total number of tested human sera for each year in which they were collected.
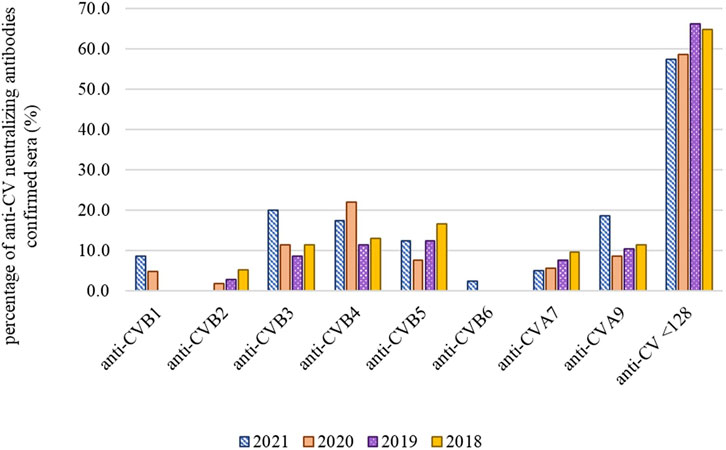
Figure 3. Percentage of anti-CV neutralizing antibodies confirmed samples from the total number of tested human sera samples for each year in which they were collected. In anti-CV samples <128, the titer of neutralizing antibodies against CVB1-6, A7 and A9 was lower than 128, antibodies against other serotypes were not tested.
Discussion
Enteroviruses are widespread globally, and several serotypes have been detected in the population. Brouwer et al. (2021) summarized 153 published articles focusing on EV seroprevalence and found that 82 EV serotypes were detected in the population, with species EV B and D being the most frequently represented in Europe. The data meta-analysis included 71 studies from 13 countries and focused on the seroprevalence of EVA71 and CVA6, which are among the most common causative agents of hand, foot, and mouth disease. In some studies, the seroprevalence of EVA71 was reported as high as 88.8%, while the results of a data meta-analysis showed 45.9% positivity. In the CVA6, the analyzed studies set the seroprevalence from 40.8% to 80.9%, while the data meta-analysis set it at 58.3% (Shi et al., 2023a). Another meta-analytic study by Shi et al. (2023b) demonstrated that EVD68 seropositivity was 76%, this study included 13 publications from China, Taiwan, Japan, Malaysia, the Netherlands, the United Kingdom, and the United States. These are just a few examples of the substantial number of EV serotypes. Since infection can occur inapparently or with non-specific mild symptoms, a person can overcome EV infection without knowing it and the prevalence in the population can be even higher.
Enzyme-linked immunosorbent assay using an appropriate antigen can be a useful tool for the detection of anti-EV antibodies against several serotypes, and because ELISA involves multiple steps, optimization is the key to success. The high prevalence of EV seropositivity is one of the two main reasons why it is difficult to find negative sera samples (or sera that do not contain any anti-EV antibodies) needed to establish a cut-off value. Another obstacle in determining the in-house ELISA cut-off is the mandatory PV vaccination, because the KTL-510 peptide synthesized based on the immunodominant region of the PV type 3 VP1 protein, which we used as an antigen in the peptide-ELISA, also detects antibodies against PV 1 to 3 (Roivainen et al., 1991; Hovi and Roivainen, 1993). In Slovakia, the first vaccination against poliomyelitis is given in the third, sixth, and eleventh months of life and the re-vaccination at the age of six and thirteen (Public Health Office of the Slovak Republic, 2024), as a result, it is possible to detect these antibodies at certain levels even in small children. All these facts explain why the set 2.04 ± 0.04 value, which may be relatively high, even though we determined the value based on samples which were previously evaluated as negative by commercial ELISA kit or VNT. The limiting factors of this comparison are that with samples evaluated for the presence of anti-EV IgG antibodies by a commercial kit, which does not describe the serotypes it captures and VNT captures only neutralizing antibodies against 8 selected CV serotypes.
There are differences in the published literature as to whether human serum inactivation is necessary for different serological methods. Some authors state that serum inactivation can cause false positive or false negative results (Lin et al., 2021). However, the data diverges between different publications in which antibodies against the same virus are detected (Hu et al., 2020a; 2020b; Sapkal et al., 2020; Xue et al., 2020; Rungrojcharoenkit et al., 2023). For detecting antibodies against different EVs from mouse and human sera, samples were inactivated at 56°C for 30 min (Huang et al., 2010; Yang et al., 2014; Hou et al., 2015; Liu et al., 2019). Bendig and Molyneaux (1996) did not inactivate patient sera in μ-capture ELISA for the detection of EV IgM. However, the mentioned studies on different EVs did not compare the results of inactivated and non-inactivated sera and therefore did not determine how it affects the results. Inactivation at 56°C for 30 min is a standard step for VNT, while it is not recommended for many commercial ELISA kits. Because the publications for individual serological methods focused on temperature inactivation were not uniform, we evaluated and compared inactivated at 56°C for 30 min and 50°C for 30 min and non-inactivated sera. The measured values of OD were highest in sera samples inactivated at 56°C and decreased with declining temperature. These results agree with published studies that detected an increase in the level of IgG antibodies after serum inactivation (Fante et al., 2021; Lin et al., 2021). As the reason for this effect, Fante et al. (2021) reported that during thermal inactivation, IgG is released from the C1q-IgG and C3d-IgG immune complexes, which increases the measured values. Lin et al. (2021) state that under conditions of heat inactivation, monomeric IgG antibody molecules can aggregate, and therefore the detection values of IgG antibodies increase. Thermal polymerization of IgG affects its biological activity and the structure of its products, with the degree of polymerization increasing with heat and the strongest binding ability being at a temperature of 62°C. The conformation of IgG changes and the number and availability of active sites of the antibody also increases. This reaction is non-specific and can result in false positivity. In our case, however, we can exclude false negativity and false positivity of samples after inactivation, because we set the cut-off value based on 100 samples inactivated at 56°C for 30 min with the lowest OD values from the entire sample set. Moreover, the majority of publications whose results were distorted after sera inactivation used serological methods other than the in-house ELISA method and focused on viruses other than EVs.
In the studied serum samples collected between 2018 and 2021, we found that in 54 cases out of 402 (13.4%) samples (dilution of 1:1,000 by an in-house ELISA method with KTL-510 peptide) the measured level of OD anti-EV IgG was higher as a set limit of 2.04. We also evaluated samples, diluted at 1:100 dilution, OD values were above the cut-off in 310 out of 402 samples (77%), while these data correspond more with the results of seroprevalence studies of Shi et al. (2023a); Shi et al. (2023b). However, we consider for the determination of cut-off values OD values obtained at 1:1,000 dilution as the authoritative data for determining the positivity of patient samples. We also assessed a set of 402 samples by VNT for the presence of anti-CV antibodies. We found that the prevalence of neutralizing antibodies to individual CV serotypes varied from year to year, which may be a sign of smaller epidemics caused by particular serotypes. In 2018, antibodies against CVB5 were the most common (16.7%), as well as in 2019, at 12.5%. In 2019, however, anti-CVB4 (11.5%) and anti-CVA9 (10.6%) neutralizing antibodies were present in a similar number of cases, and the total number of positive cases was lower compared to other monitored years. In 2020, the most represented were anti-CVB4 neutralizing antibodies, and in 2021, anti-CVB3 (20%), anti-CVA9 (18.8%) and anti-CVB4 (17.5%). The results of VNT and in-house ELISA are not comparable because we used different sample dilutions for both methods. Using VNT, we detected neutralizing antibodies against eight selected CV serotypes, whereas using ELISA, we detected only IgG, but against a larger number of different EV serotypes. The big advantage of ELISA is that it saves time and the examination of a larger number of samples is done within 1 day (one run), unlike VNT where the results are obtained within a week.
In conclusion, we can say that the optimized in-house ELISA is suitable for routine diagnosis of patient samples, as it is timesaving. It captures a wide range of EV serotypes, PV type 1–3, CVA9, A13, A16, A18, A21, CVB1 - B6, 23, ECV1 - 9, 11, 13–15, 17–19, and 21, and EV69, EV70, and EV71 (according to study of Hovi and Roivainen, 1993). It is a good supporting method for the diagnosis of viral infections. It may be an useful tool in different applications ranging from serological surveillance studies to vaccine or antiviral treatment effectivity examinations.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies involving humans because the samples used in the studies were sent to the National Reference Center (NRC) laboratories and Office of Public Health (OPH) laboratories. They were used retrospectively. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from NRC, OPH as mentioned above. The samples are part of epidemiological surveillance programs. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
Conceptualization: JG and SB; methodology: MP, VS, KA, and ST; writing-original draft preparation: MP; writing, review, and editing: VS, KA, BB, MB, SB, and ST. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Reference Center for Identification of Enteric Viruses and Slovak Medical University internal grant number 05/2021-SVG1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2024.12739/full#supplementary-material
Abbreviations
CV, coxsackievirus; ECV, echovirus; ELISA, enzyme-linked immunosorbent assay; EV, enterovirus; Ig, immunoglobulin; OD, optical density; PBS, phosphate buffered saline; PCR, polymerase chain reaction; PFTM, PBS with 1% fetal bovine serum, 1% bovine serum albumin, 0.1% Tween 20 and 5 mM ethylenediaminetetraacetic acid; PV, poliovirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation; VNT, virus neutralization test; VP, viral protein.
References
Bendig, J. W., and Molyneaux, P. (1996). Sensitivity and specificity of mu-capture ELISA for detection of enterovirus IgM. J. Virol. Methods 59, 23–32. doi:10.1016/0166-0934(95)01997-9
Brouwer, L., Moreni, G., Wolthers, K. C., and Pajkrt, D. (2021). World-wide prevalence and genotype distribution of enteroviruses. Viruses 13, 434. doi:10.3390/v13030434
Cao, Y., Li, K., Xing, X., Zhu, G., Fu, Y., Bao, H., et al. (2022). Development and validation of a competitive ELISA based on bovine monoclonal antibodies for the detection of neutralizing antibodies against foot-and-mouth disease virus serotype A. J. Clin. Microbiol. 60, e0214221. doi:10.1128/jcm.02142-21
Chapman, N. M., and Kim, K. S. (2008). Persistent coxsackievirus infection: Enterovirus persistence in chronic myocarditis and dilated cardiomyopathy. Curr. Top. Microbiol. Immunol. 323, 275–292. doi:10.1007/978-3-540-75546-3_13
Craig, M. E., Robertson, P., Howard, N. J., Silink, M., and Rawlinson, W. D. (2003). Diagnosis of enterovirus infection by genus-specific PCR and enzyme-linked immunosorbent assays. J. Clin. Microbiol. 41, 841–844. doi:10.1128/JCM.41.2.841-844.2003
Fante, M. A., Decking, S. M., Bruss, C., Schreml, S., Siska, P. J., Kreutz, M., et al. (2021). Heat-inactivation of human serum destroys C1 inhibitor, pro-motes immune complex formation, and improves human T cell function. Int. J. Mol. Sci. 22, 2646. doi:10.3390/ijms22052646
Hochman, O., Xu, W., Yang, M., Yang, C., Ambagala, A., Rogiewicz, A., et al. (2023). Development and validation of competitive ELISA for detection of H5 hemagglutinin antibodies. Poultry 2, 349–362. doi:10.3390/poultry2030026
Hou, W., Yang, L., He, D., Zheng, J., Xu, L., Liu, J., et al. (2015). Development of a coxsackievirus A16 neutralization test based on the enzyme-linked immunospot assay. J. Virol. Methods 215-216, 56–60. doi:10.1016/j.jviromet.2015.02.010
Hovi, T., and Roivainen, M. (1993). Peptide antisera targeted to a conserved sequence in poliovirus capsid VP1 cross-react widely with members of the genus Enterovirus. J. Clin. Microbiol. 31, 1083–1087. doi:10.1128/jcm.31.5.1083-1087.1993
Hu, X., An, T., Situ, B., Hu, Y., Ou, Z., Li, Q., et al. (2020a). Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. J. Clin. Lab. Anal. 34, e23411. doi:10.1002/jcla.23411
Hu, X., Zhang, R., An, T., Li, Q., Situ, B., Ou, Z., et al. (2020b). Impact of heat-inactivation on the detection of SARS-CoV-2 IgM and IgG antibody by ELISA. Clin. Chim. Acta. 509, 288–292. doi:10.1016/j.cca.2020.06.032
Huang, M. L., Chiang, P. S., Luo, S. T., Liou, G. Y., and Lee, M. S. (2010). Development of a high-throughput assay for measuring serum neutralizing antibody against enterovirus 71. J. Virol. Methods 165, 42–45. doi:10.1016/j.jviromet.2009.12.015
Hubert, F. (2018). A logical introduction to probability and induction. New York: Oxford University Press.
International Committee on Taxonomy of Viruses (2023). Enterovirus taxonomy. Available at: https://ictv.global/report/chapter/picornaviridae/picornaviridae/enterovirus (Accessed November 27, 2023).
Itani, T., Chalapa, V., Semenov, A., and Sergeev, A. (2023). Laboratory diagnosis of nonpolio enteroviruses: a review of the current literature. Biosaf. Health 5, 112–119. doi:10.1016/j.bsheal.2022.12.002
Lin, J., Dai, W., Li, W., Xiao, L., Luo, T., Guo, Y., et al. (2021). Potential false-positive and false-negative results for COVID-19 IgG/IgM antibody testing after heat-inactivation. Front. Med. 7, 589080. doi:10.3389/fmed.2020.589080
Liu, D., Xu, L., Zhu, R., Yin, Z., Lin, Y., Hou, W., et al. (2019). Development of an efficient neutralization assay for Coxsackievirus A10. Appl. Microbiol. Biotech. 103, 1931–1938. doi:10.1007/s00253-018-09598-7
Murray, A., and Lawrence, G. P. (1993). How should the repeatability of clinical measurements be analysed? An assessment of analysis techniques with data from cardiovascular autonomic function tests. Q. J. Med. 86, 831–836.
Oluka, G. K., Namubiru, P., Kato, L., Ankunda, V., Gombe, B., Cotten, M., et al. (2023). Optimisation and validation of a conventional ELISA and cut-offs for detecting and quantifying anti-SARS-CoV-2 spike, RBD, and nucleoprotein IgG, IgM, and IgA antibodies in Uganda. Front. Immunol. 14, 1113194. doi:10.3389/fimmu.2023.1113194
Pallansch, M. A., Oberste, M. S., and Whitton, J. L. (2013). “Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses,” in Fields virology. Editors D. M. Knipe, and P. M. Howley 6th ed. (Philadelphia, PA: Lippincott Williams & Wilkins), 490–530. Volume 1.
Payne, S. (2017). “Chapter 4 - methods to study viruses,” in Viruses. Editor S. Payne (Cambridge, MA: Academic Press), 37–52.
Public Health Office of the Slovak Republic (2024). Vaccination calendar. Available at: https://www.uvzsr.sk/documents/d/uvz/ockovaci-kalendar-na-rok-2024-povinne-pravidelne-ockovanie-deti-a-dospelych-pdf (Accessed January 3, 2024).
Roivainen, M., Alfthan, G., Jousilahti, P., Kimpimäki, M., Hovi, T., and Tuomilehto, J. (1998). Enterovirus infections as a possible risk factor for myocardial infarction. Circulation 98, 2534–2537. doi:10.1161/01.cir.98.23.2534
Roivainen, M., Närvänen, A., Korkolainen, M., Huhtala, M. L., and Hovi, T. (1991). Antigenic regions of poliovirus type 3/sabin capsid proteins recognized by human sera in the peptide scanning technique. Virology 180, 99–107. doi:10.1016/0042-6822(91)90013-2
Rungrojcharoenkit, K., Suthangkornkul, R., Utennam, D., Buddhari, D., Pinpaiboon, S., Mongkolsirichaikul, D., et al. (2023). Standardization of in-house anti-IgG and IgA ELISAs for the detection of COVID-19. PLoS One 18, e0287107. doi:10.1371/journal.pone.0287107
Sapkal, G., Shete-Aich, A., Jain, R., Yadav, P. D., Sarkale, P., Lakra, R., et al. (2020). Development of indigenous IgG ELISA for the detection of anti-SARS-CoV-2 IgG. Indian J. Med. Res. 151, 444–449. doi:10.4103/ijmr.IJMR_2232_20
Shi, Y., Chen, P., Bai, Y., Xu, X., and Liu, Y. (2023a). Seroprevalence of coxsackievirus A6 and enterovirus A71 infection in humans: a systematic review and meta-analysis. Arch. Virol. 168, 37. doi:10.1007/s00705-022-05642-0
Shi, Y., Ran, Q., Wang, X., and Shi, L. (2023b). Seroprevalence of enterovirus D68 infection among humans: a systematic review and meta-analysis. Intervirology 66, 111–121. doi:10.1159/000531853
Smatti, M. K., Cyprian, F. S., Nasrallah, G. K., Al Thani, A. A., Almishal, R. O., and Yassine, H. M. (2019). Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses 11, 762. doi:10.3390/v11080762
Swanink, C. M., Veenstra, L., Poort, Y. A., Kaan, J. A., and Galama, J. M. (1993). Coxsackievirus B1-based antibody-capture enzyme-linked immunosorbent assay for detection of immunoglobulin G (IgG), IgM, and IgA with broad specificity for enteroviruses. J. Clin. Microbiol. 31, 3240–3246. doi:10.1128/jcm.31.12.3240-3246.1993
Xue, X., Zhu, C., Huang, S., Pan, L., Xu, J., and Li, W. (2020). Effect of heat inactivation of blood samples on the efficacy of three detection methods of SARS-CoV-2 antibodies. J. South. Med. 40, 316–320. doi:10.12122/j.issn.1673-4254.2020.03.03
Keywords: serology, ELISA, antibodies, enterovirus, optimization
Citation: Pellerova M, Albertova K, Simkova V, Borsanyiova M, Benkoova B, Kissova R, Pastuchova K, Tauriainen S, Galama JMD and Bopegamage S (2024) Detection of anti-enterovirus IgG in human sera by ELISA method using the KTL-510 peptide. Acta Virol. 68:12739. doi: 10.3389/av.2024.12739
Received: 25 January 2024; Accepted: 13 September 2024;
Published: 25 September 2024.
Edited by:
Katarina Polcicova, Slovak Academy of Sciences, SlovakiaReviewed by:
Malyaj R. Prajapati, Indian Agricultural Research Institute (ICAR), IndiaEva Varečková, Slovak Academy of Sciences, Slovakia
Copyright © 2024 Pellerova, Albertova, Simkova, Borsanyiova, Benkoova, Kissova, Pastuchova, Tauriainen, Galama and Bopegamage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michaela Pellerova, bWljaGFlbGEucG9zcGlzaWxvdmFAc3p1LnNr
 Michaela Pellerova
Michaela Pellerova Katarina Albertova1
Katarina Albertova1 Maria Borsanyiova
Maria Borsanyiova Brigita Benkoova
Brigita Benkoova Shubhada Bopegamage
Shubhada Bopegamage