Abstract
Puumala orthohantavirus (PUUV) is a causative agent of haemorrhagic fever with renal syndrome, mainly transmitted through rodent excretions. Classification of PUUV has traditionally been based on the geographic locations of their discovery, but this system faces challenges due to inconsistencies between the names of lineages and their actual distribution. In this study, we propose a quantitative criterion of “10% nucleotide difference” to distinguish PUUV genotypes, resulting in a subdivision of PUUV into twenty-one distinct genotypes. The application of this criterion suggested a further division of the CE, RUS, FIN and N-SCA lineages and was consistent with the current taxonomic position of the S-SCA, LAT, DAN and ALAD lineages. In addition, analysis of reassortment and recombination events showed that PUUV undergoes these processes, but they are relatively rare. Our results emphasise the importance of considering genetic distances in the classification of PUUV and provide a robust criterion for subspecies-level taxonomy.
Introduction
Puumala orthohantavirus (PUUV) is a species included in the genus Orthohantavirus, family Hantaviridae and order Bunyavirales. Hantaviruses have a negative-sense single-stranded RNA genome divided into three segments: small (S), medium (M), and large (L), which are represented by three RNA molecules. These segments encode nucleocapsid protein (N), glycoprotein precursor (G) that matures into the surface glycoproteins Gn and Gc (Cifuentes-Muñoz et al., 2014), and RNA-dependent RNA polymerase (RdRp), respectively (Plyusnin et al., 1996). In addition, Puumala S segment encodes a small non-structural protein (NSs) by the overlapping open reading frame (Jääskeläinen et al., 2007).
Puumala orthohantavirus causes nephropathia epidemica, a mild form of hemorrhagic fever with renal syndrome (HFRS). The course of HFRS is highly variable, ranging from frequently asymptomatic to a lethal outcome. The most common symptoms are high fever, headache, abdominal pains, backache and nausea or vomiting (Vaheri et al., 2013). Humans can be infected through animal bites or by inhaling aerosols from rodent excreta (Blinova et al., 2022). However, transmission of PUUV via transfusion of platelets or other blood products has been documented (Sinisalo et al., 2010). Hantaviral species are strongly associated with its host. Bank voles (Myodes glareolus) are a reservoir of Puumala virus. These rodents are widespread in the vast territories in Europe and Western Siberia, where PUUV has been found (Davidyuk et al., 2020).
Actual PUUV classification
To date, there is no standardised classification of the subspecies of Puumala virus based on quantitative criteria. The current classification consists of several lineages such as North-Scandinavian (N-SCA), Finnish (FIN), Russian (RUS), Latvian (LAT), Alpe-Adrian (ALAD) and Central European (CE) (Sironen et al., 2001; Dekonenko et al., 2003; Castel et al., 2019; Blinova et al., 2022; 2023). Some sequences from the same lineage may be genetically more distant from each other than sequences from different lineages, as there are currently no strict thresholds for the determination of the lineages. Furthermore, some new sequences cannot be clearly assigned to one of the current lineages (Blinova et al., 2023). Here, we propose a threshold for genetic distance, to separate the lineages unambiguously. On the other hand, we show that such a quantitative criterion can be used to divide PUUV into distinct genotypes.
Puumala recombination and reassortment
Phylogenetic reconstruction is only reliable for sequences without horizontal gene transfer, and for viruses, this means that only parts of the genome that do not undergo recombination can be used. Recombination occurs when at least two viruses co-infect the same host cell and exchange genetic segments. Different types of viral recombination are distinguished by the structure of the crossover site. Homologous recombination occurs at the same site in both parental strands, whereas non-homologous or illegitimate recombination occurs at different sites of the genetic fragments involved. Reassortment occurs in viruses with segmented genomes that can exchange complete segments (Pérez-Losada et al., 2015). Since PUUV has three segments, it is important to understand whether its genome undergoes reassortment and homologous recombination before conducting any studies on its phylogeny.
Homologous recombination is an important aspect of viral evolution in the context of phylogenetic analysis, as different parts of the genome can evolve independently. Since Puumala viruses have three RNA molecules in their virion they can also exchange whole genome segments.
Homologous recombination has been reported for hantaviruses. E.g., it occurs in Tula virus and Dobrava-Belgrade virus which are the relatives of PUUV (Sibold et al., 1999; Klempa et al., 2003). In addition, numerous reassortment events have been found in other hantaviruses (Kim et al., 2016; Lee et al., 2017). Due to their host specificity, there can only be one species of hantavirus in a host species, so that hantaviruses, unlike other bunyaviruses, can generally only undergo reassortment events within the same viral species (Klempa, 2018). Reassortment is well described for Puumala orthohantavirus. In the studies in Finland, a high frequency of reassortment, 19.1%–32%, was observed. The reassortment events within Finnish lineage, between Finnish and North Scandinavian groups have been found (Razzauti et al., 2008; 2009). The authors estimated the contributions of homologous recombination and reassortment and concluded that recombination is rarer than reassortment. In PUUV from Central Europe, sequences from Slovakia were clustered with sequences from Bohemia (Czech Republic) and Bavaria (Germany) only for M, but not for S segments. This means that the M segments of these viruses are closely related, but the S segments are not. The S segment of the Slovakian viruses was related to the S segments of PUUV from Belgium and France. This fact may indicate reassortment events (Razzauti et al., 2009). On the other hand, this study was performed for partial genomes, so these observations can also be explained by the presence of homologous recombination.
The presence of reassortment events means that the phylogenies for the different segments do not match. At the same time, different regions within the same segment may also have different evolutionary histories. Therefore, the segment without signs of recombination should be used for phylogenetic analysis. On the other hand, the segments are not evenly sequenced. Using the most frequently sequenced segments makes it possible to comprehensively uncover the phylogeny of PUUV.
Materials and methods
Recombination and reassortment analysis
Only viruses with three fully sequenced segments were selected for recombination and reassortment analysis (total number of sequences = 50). The GenBank database was searched for viruses for which all three segments were available. Then these genomic sequences were joined together with strings of 700 N’s between segments for each PUUV. In addition, only the protein-coding regions were used for the analysis. These sequences were aligned using the Muscle algorithm in MEGA software (Kumar et al., 2018) and then recombination and reassortment events were searched for using the online web application Shiny1 (Vakulenko et al., 2021). For each pair of genomic regions, the distances between all possible pairs of sequences were calculated and then plotted on a pairwise nucleotide distance comparison plot (PDCP) (Vakulenko et al., 2021). If no recombination or reassortment has taken place, the distance between two sequences in one region should correlate with the distance in another region. If recombination or reassortment has occurred, these distances may not correlate. Without recombination and reassortment, the distances between pairs of sequences for each pair of genomic regions should follow a linear relationship, so a high value of root mean square error (RMSE) indicates recombination or reassortment. A pairwise distance deviation matrix (PDDM) was then calculated for all possible pairs of genomic regions using a sliding window with a length of 700 nt and a step of 50 nt. The PDDM thus summarises the RMSE of all PDCPs. Red areas on the PDDM indicate a high RMSE value and thus the fact that the corresponding genome regions could originate from different ancestors. Blue areas indicate a lower probability of recombination or reassortment.
To detect recombination and reassortment event GiRaF (Nagarajan and Kingsford, 2011) and RDP4 (Martin et al., 2015) software was also implemented. GiRaf used as an input phylogenetic trees that were obtained using Bayesian algorithms by the MrBayes software (Huelsenbeck and Ronquist, 2001). In RDP4 seven methods were used to find possible recombination and reassortment events (RDP, GENECONV, Chimaera, MaxChi, BootScan, SiScan, and 3Seq). When looking for potential recombination inside each one of the segments with RDP4, we used alignments of sequences for that particular segment, and when analyzing reassortment events between different segments, we used a full-genome alignment with segments separated by N’s, as mentioned before.
The visualisation of the similarity was created using the SimPlot software (Lole et al., 1999). It shows the comparison between a set of sequences and a reference sequence.
The trees for the recombination and reassortment analysis were created using the maximum likelihood algorithm with 100 bootstraps with the MEGA software and visualised with the Figtree software (Rambaut, 2020).
Phylogenetic analysis
All 514 currently available (as of June 30th, 2023) Puumala S segment sequences were obtained from the GenBank. Only sequences that contain a complete nucleocapsid coding fragment were used in the study (n = 505). A phylogenetic tree was built using the maximum likelihood method and branches were named according to currently accepted systematics.
Protein and nucleotide pairwise distances were calculated using MEGA software. Scatter plots in nucleotide distances/protein distances coordinates were built using R Studio [ggplot2 library (Wickham, 2016)]. CD-HIT (Fu et al., 2012) was used for sequence clustering to visualise the results obtained by rationally reducing the sample size. Then another maximum likelihood phylogenetic tree was built from the clustered sequences using IQ-TREE with ultrafast bootstrap analysis (n = 1,000) (Nguyen et al., 2015). The protein and nucleotide sequences are available at2.
Results
Tentative quantitative criteria for the PUUV taxonomic assignment at the subspecies level
The S segment was the most represented in the GenBank segment (as of June 30th, 2023 number of sequences = 514) since it has the shortest length and it’s sequencing is the most cost-effective. L segment was the least represented segment (number of sequences = 65). RDP4 software was used to determine whether the S segment undergoes recombination events. No event was found that would have been supported by more than two methods out of the seven used.
Since sequences of other segments are not available for most viruses and the S segment doesn’t seem to undergo any recombination events, the phylogenetic analysis was based on the nucleocapsid protein gene located in the S segment (n = 505). The distributions of pairwise nucleotide and amino acid distances were plotted to visualize the sequence space occupied by PUUV (Figure 1). Each pair of sequences in a dataset was represented by a dot with coordinates reflecting the nucleotide and amino acid sequence distance between them. It should be noted that without strict quantitative criteria, the taxonomic classification may be ambiguous and may change as new distinct sequences are discovered. Each dot was coloured according to whether the virus pair consisted of inter- or intragroup viruses. The current classification of PUUV lineages was used in Figure 1A to define the inter- or intralineage assignment of the virus pairs (Figure 1A). At the same time, PUUVs were clustered based on the threshold of 10% of nucleotide sequences. These groups of sequences were used for the inter- or intracluster assignment of the virus pair in Figure 1B.
FIGURE 1
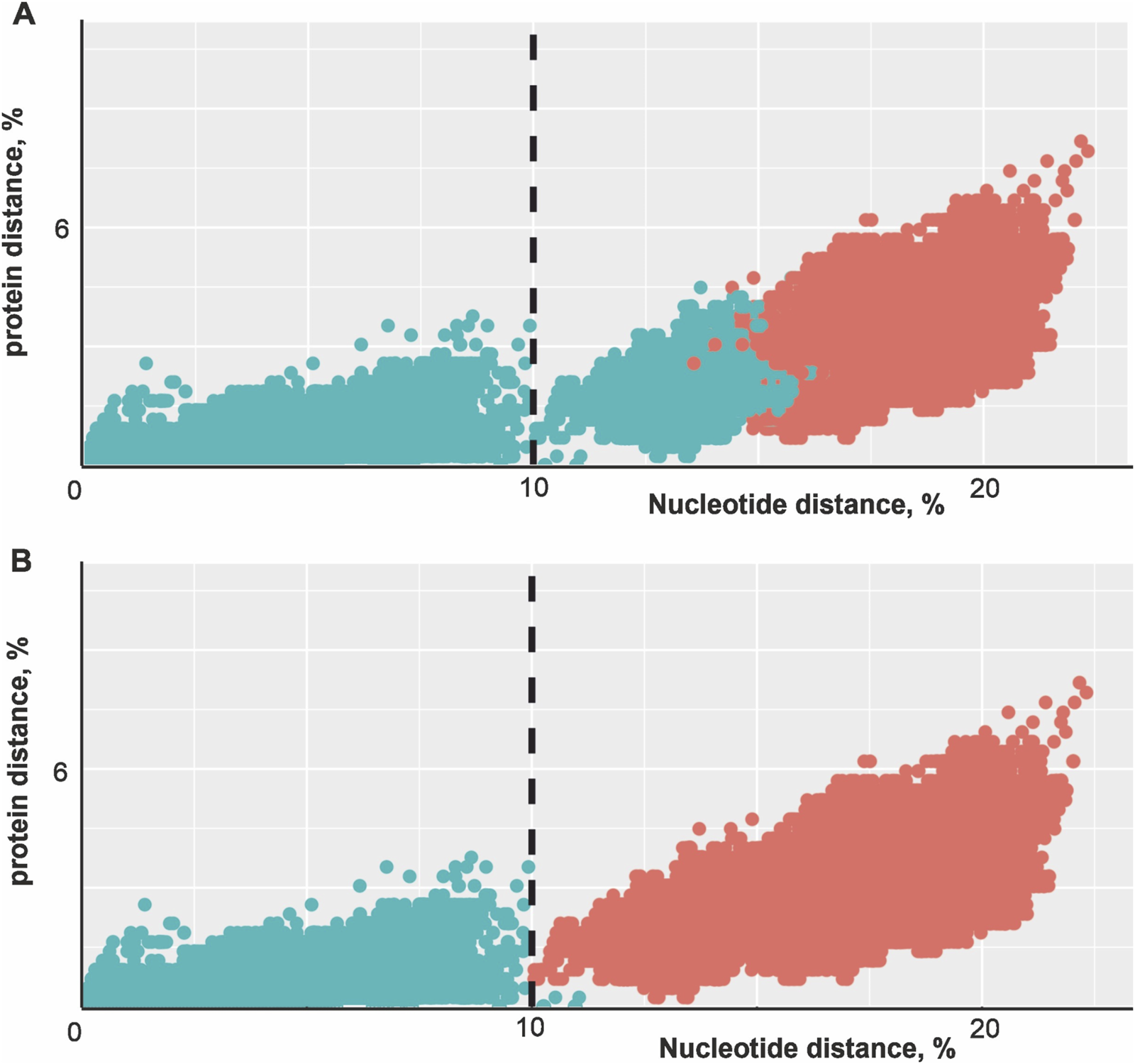
Scatter plot of amino acid and nucleotide distances for pairs of PUUVs S segments. (A) Intralineage virus pairs were indicated by cyan dots, interlineage virus pairs by red dots. Lineage assignments were made according to (Castel et al., 2019; Blinova et al., 2023). The threshold of 10% nucleotide difference is shown as a vertical dashed line. (B) Implementation of tentative quantitative criteria for the PUUV taxonomic assignment at the subspecies level. Intracluster virus pairs (difference less than 10% nucleotide distance) are indicated by cyan dots, intercluster (difference more than 10% nucleotide distance) virus pairs by red dots. The threshold of 10% nucleotide difference is indicated as a vertical dashed line.
Two different groups of virus pairs on the plot could be clearly distinguished (Figure 1). One group of virus pairs has less than 10% nucleotide difference in the S segment sequence, while the other group differed in more than 10% of the nucleotides in the S segment sequence. Viruses from different groups always differed in more than 10% of the nucleotides, while some sequence pairs from the same group also differed by more than 10% of nucleotides (Figure 1A, cyan dots to the right of the threshold line). Indeed, sequence pairs from different lineages can be phylogenetically closer than a pair from one group. For example, #KX815394 (LAT lineage) and #JQ319167 (FIN lineage) are more similar (12.97% of nucleotides were different) than #MG812440 (CE lineage) and #AF294652 (CE lineage) (16.5% of nucleotides were different). Thus, in some cases, intralineage diversity is higher than interlineage diversity.
To test the validity of the “10% nucleotide difference” criterion usage as a quantitative measure for classifying Puumala virus, we divided the viruses into 21 groups (Figure 2). Remarkably, several lineages (CE, RUS, N-SCA, and FIN) were subdivided into multiple groups specific to the isolated regions according to this criterion (Table 1). At the same time, other lineages (ALAD, S-SCA, and DAN) correspond to the novel taxon. This means that the nucleotide distance between the coding regions of the S segments of two sequences can be the criterion for an unambiguous classification of PUUV into the subspecies taxon. At the same time, such grouping of PUUVs has its limitations (cyan dots to the right of the “10% nucleotide difference” criterion in Figure 1B). #JN657228 (LAT lineage) has a nucleotide difference of less than 10% with the sequences MT580935 and KX815394, but more than 10% with other representatives of the LAT lineage.
FIGURE 2
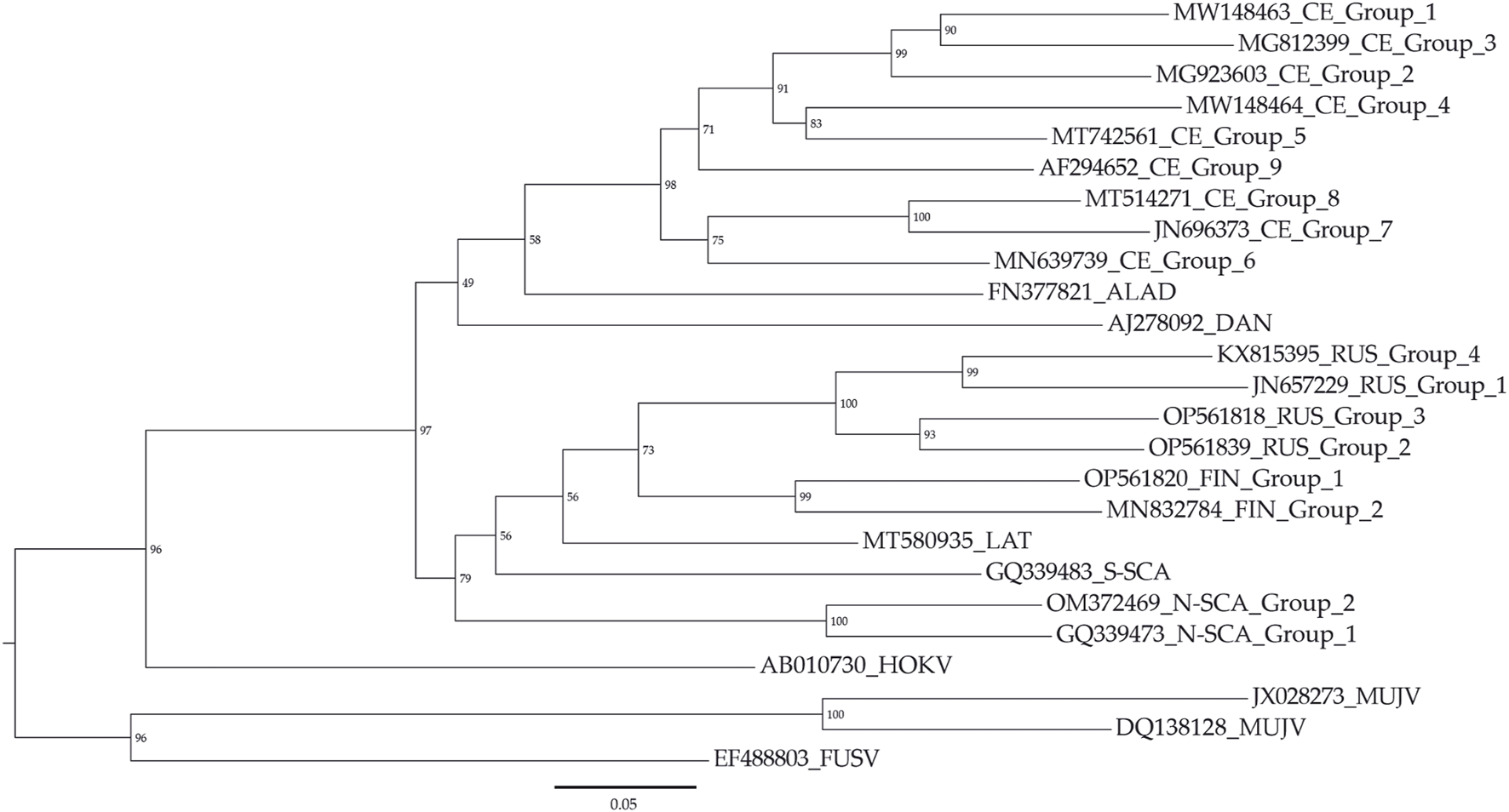
Maximum likelihood phylogenetic tree for PUUV groups. The threshold “10% nucleotide distance” was used as a criterion for PUUV clustering. Muju virus (MUJV), Hokkaido virus (HOKV), Fusong virus (FUSV) were used as outgroups.
TABLE 1
| Genotype name | Region of origin |
|---|---|
| CE group 1 | France (Ardennes, Hauts-de-France, Île-de-France, Grand Est), Germany (North Rhine-Westphalia), Belgium, Netherlands |
| CE group 2 | France (Grand Est, Hauts-de-France) |
| CE group 3 | Belgium |
| CE group 4 | France (Centre-Val de Loire, Île-de-France, Bourgogne-Franche-Comté) |
| CE group 5 | France (Grand Est, Bourgogne-Franche-Comté, Auvergne-Rhône-Alpes, Hauts-de-France), Germany (Baden-Württemberg) |
| CE group 6 | Germany (Lower Saxony), Netherlands |
| CE group 7 | Germany (Bavaria) |
| CE group 8 | Germany (Baden-Württemberg, Bavaria) |
| CE group 9 | Slovakia |
| RUS group 1 | Latvia |
| RUS group 2 | Russia (Ivanovo, Moscow, Kursk regions) |
| RUS group 3 | Russia (Saratov, Penza, Ulyanovsk, Samara regions, Udmurt, Tatarstan, Mordovia, Bashkortostan republics) |
| RUS group 4 | Poland (Subcarpathian Voivodeship) |
| FIN group 1 | Russia (Arkhangelsk, Tyumen, Omsk regions) |
| FIN group 2 | Finland (Kainuu, Central Finland, South Savo, Lapland, Kanta-Häme), Russia (Karelia republic) |
| N-SCA group 1 | Sweden (Norrbotten County) |
| N-SCA group 2 | Sweden (Västerbotten County) |
| S-SCA | Sweden (Gävleborg, Västmanland County) |
| DAN | Denmark |
| ALAD | Hungary, Austria, Balkan |
| LAT | Latvia, Lithuania, Poland (Warmian–Masurian Voivodeship) |
Geographical distribution of the designated groups of PUUVs (as stated in the GenBank).
The accession numbers of all S segment PUUV sequences assigned as “lineages” and “genotypes” are provided in Supplementary Table S1.
Reassortment analysis
The concatenated sequences of protein-coding regions of all three segments were used (number of sequences = 50). The results of reassortment and recombination analysis are shown on the pairwise distance deviation matrix (PDDM) in Figure 3. The coordinates of three genes in the final alignment with separations were: 1–1,302 for N gene, 2003–5,449 for G gene and 6,150–12620 for RdRp gene3.
FIGURE 3
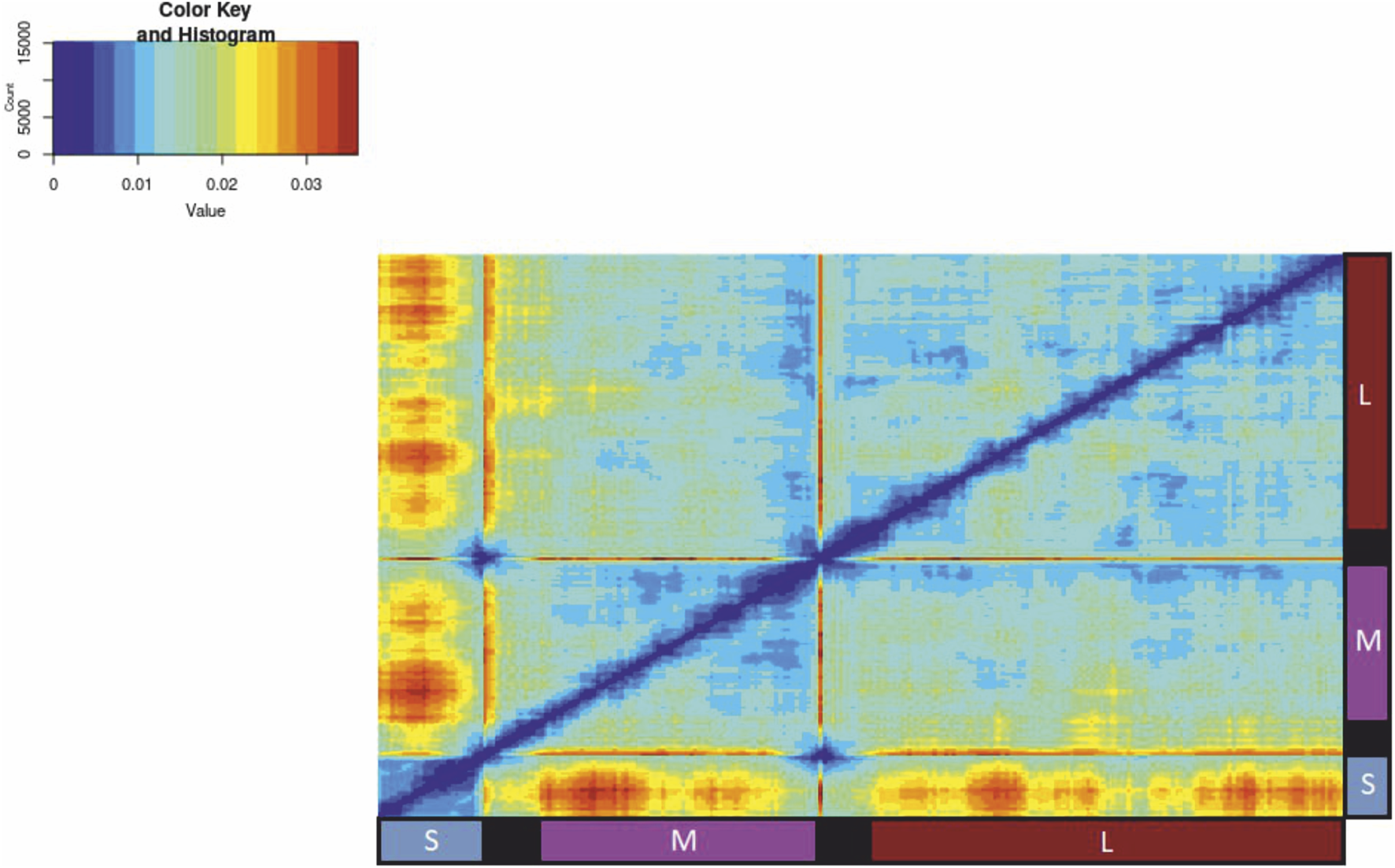
A pairwise distance deviation matrix (PDDM) with sliding window length 700 nt and 50 nt step for PUUV alignment (50 concatenated sequences of protein-coding regions of all three segments with 700 N’s between each segment for better separation). Red areas indicate a high level of sequence distance inconsistency suggesting possible presence of recombination and reassortment events.
This plot uses red colour to mark areas of high phylogenetic inconsistency among the genomic regions, while blue indicates low phylogenetic inconsistency. A high RMSE value suggests potential reassortment among the most divergent viruses in the respective genomic regions.
For the next part of the analysis, an alignment without the separation between the segments was used4. We visualised PDCPs for the following pairs of regions: S segment (1–1,302 nt in alignment) against the M segment (1,303–4,752 nt in alignment) (Figure 4B) and M segment (1,303–4,752 nt in alignment) against L segment (4,753–11,223 nt in alignment) (Figure 4C). Each sequence pair in a dataset was represented by a dot with coordinates reflecting the nucleotide sequence distance between the different genomic fragments. The density of possible distance values was indicated by colour. The PDCP for the S segment and the M segment showed that some viral pairs had phylogenetic incongruence (Figure 4B, circled) that could be explained by a reassortment event. Indeed, the S segments of these viruses differed in about 6% of the nucleotides in the S segment and in about 14% of the nucleotides in the M segment. To obtain a negative control, we concatenated the S segment and the M segment, selected odd or even sites from this artificial sequence and calculated the pairwise distances for this data set (Figure 4A).
FIGURE 4
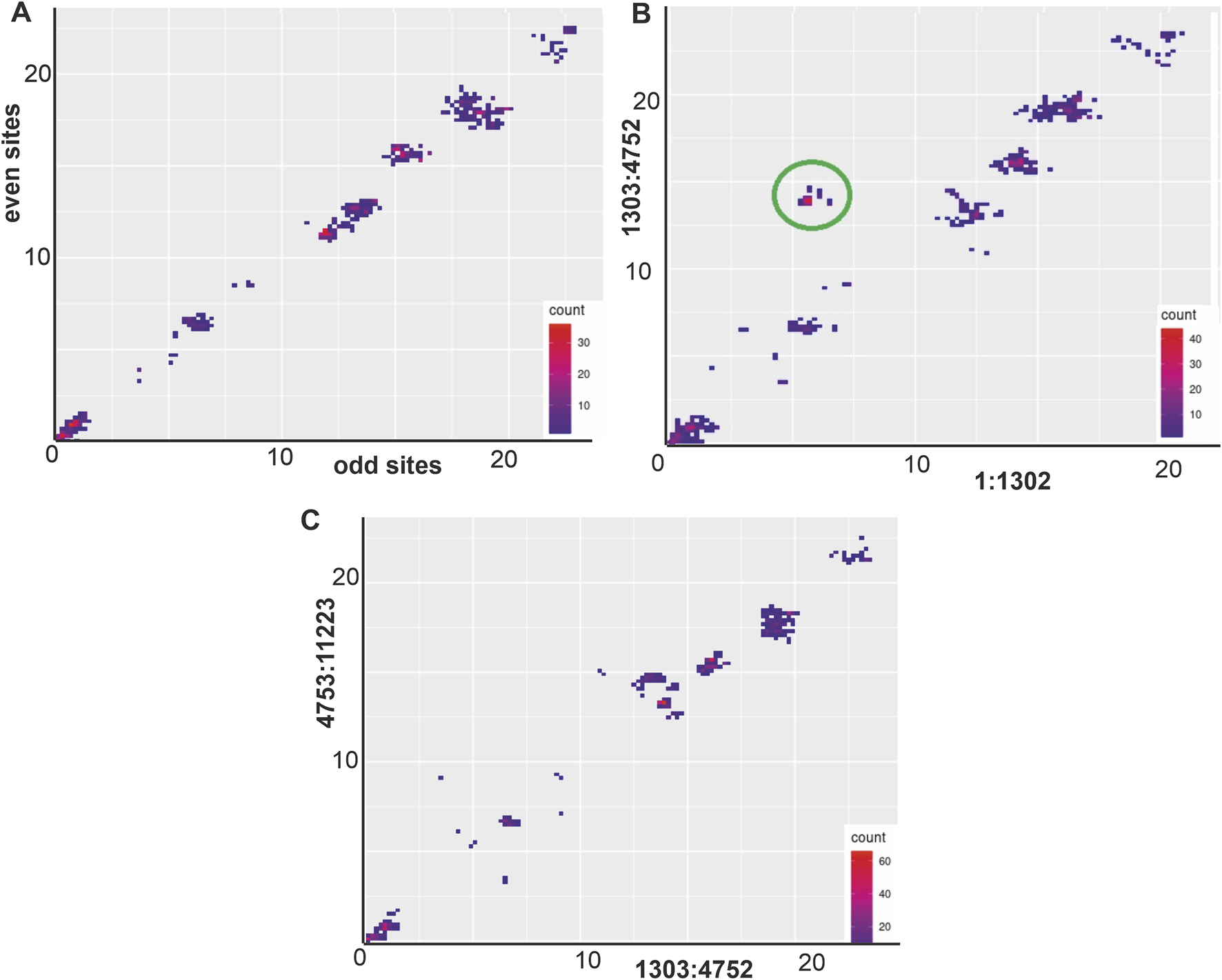
(A) PDCP control plot between odd and even positions of S segment and M segment (1–4,752 nt in alignment). (B) PDCP between the S segment (1–1,302 nt in alignment) and M segment (1,303–4,752 nt in alignment). (C) PDCP between M segment (1,303–4,752 nt in alignment) and L segment (4,753–11,223 nt in alignment).
Two concatenated viral genomic sequences that were off trend (Figure 4B, circled): “strain K27” (#MZ673552-MZ673554) and “isolate 989” (#OL343606, OL343584, and OL343562) were selected for further analysis. The similarity plot for this pair of sequences and three control sequences: “isolate Klyukva” (#MZ580951-MZ580953), “strain Osnabruck V29” (#MN639737-MN639739) and Muju virus “isolate 11-1” (#JX028271-JX028273) was shown in Figure 5A. The sequence of “isolate 989” was used as a reference. Phylogenetic trees of these five sequences were constructed using the S segment and the M segment (Figure 5B). Both trees showed high bootstraps but different topologies. The S segment of the sequence “isolate 989” was close to the S segment of the sequence “strain K27,” while the M segment of these viruses was more diverse.
FIGURE 5
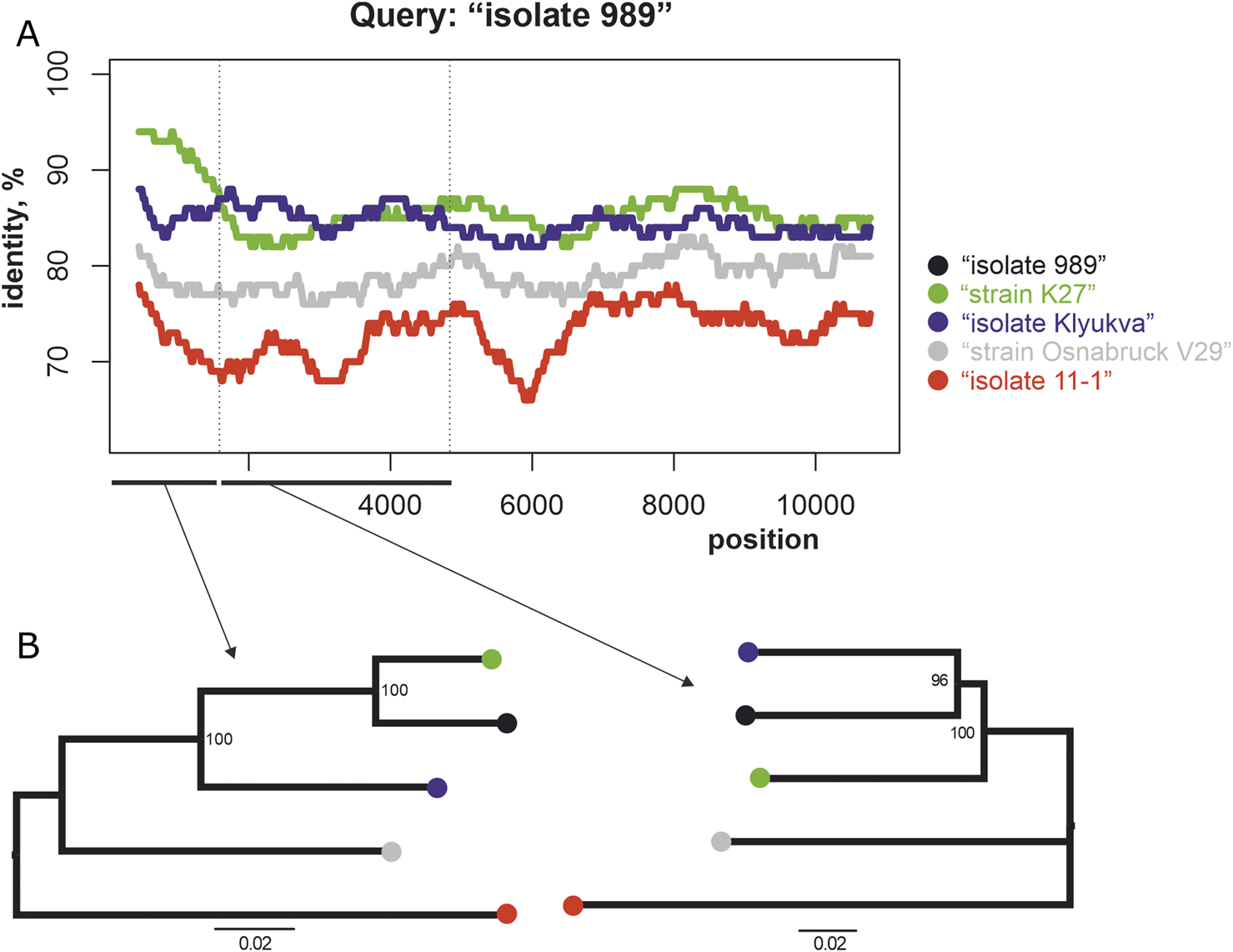
(A) Similarity plot for the set of sequences. Each point is the percentage of identity compared to the concatenated genomic sequences of “isolate 989” (#OL343606, OL343584, and OL343562) within the 900 bp wide sliding window with the 20 bp step. (B) The phylogenetic trees for these sequences show different topologies for the S (left) and the M (right) segments.
The S segment of strain K27 has 94.5% identical nucleotides with the segment of isolate 989, while its M and L segments have 86.9% and 87.4% identical nucleotides respectively. The most plausible explanation for this fact is a reassortment event in which the S segment was transmitted from a precursor virus whose descending M and L segments are currently unknown. However, this is speculation, and without direct evidence, the drastic change in the proportion of identical nucleotides in the different segments can also be explained by the regional differences in mutation rates. According to the similarity plot, the different regions of the M segment have varying percentages of identical nucleotides (Figure 5A), and the phylogenetic trees of these regions also have different topologies5. In addition, conflicting tree topologies were observed for the phylogenetic trees for these M-segment fragments generated for all PUUVs from the dataset (6, red arrows). These observations can be explained by possible recombination events in the M-segment.
In addition, GiRaF and RDP4 were used. GiRaF found no potential reassortment events, and RDP4 confirmed the reassortment event described in this study with three of the seven methods used7. To further validate the results, phylogenetic trees for each segment were built. Preliminarily all of the segments were analyzed by RDP4 for possible recombination events. Recombinant sequence “isolate 836” (#OL343604, OL343582, OL343560) was omitted from the analysis. The phylogenetic trees8 for the S segment and the L segment have the same topology for the clades containing sequences “isolate 989,” “K27” and “Klyukva” that were previously mentioned, but for the M segment the topology is different (red arrows on the phylogenetic tree). Another potential reassortment event was discovered during the same analysis between clades that contain the sequences “Suo/renal,” “Sotkamo,” and “Mg7/2008,” respectively (blue arrows). The topologies for the S and M segment are identical for those clades, and the one for the L segment is different from them. Interestingly, an event between the same clades was discovered by RDP4 as well, although it suggests that the M segment reassorts, not L.
Discussion
Previous studies (Razzauti et al., 2008; 2009; Szabó et al., 2017) have highlighted the occurrence of reassortments and recombinations within the evolution of Puumala orthohantaviruses (PUUVs). Our study is consistent with these conclusions (Figures 3–5). Nevertheless, reassortment and recombination are relatively rare events - 162 of 1,225 sequence pairs in the PUUV dataset (50 concatenated complete protein-coding genomic regions) showed evidence of a mixed origin according to the RMSE method (circled in Figure 4). The S segments of some sequences clustered with one virus, while their M and L segments grouped with another virus (Figure 5). This was confirmed by phylogenetic trees constructed using the S segment and the M segment, which had different topologies (Figure 5). The S segment of “isolate 989,” a representative of RUS group 3, has 94% identical nucleotides with the S segment of “strain K27,” another representative of RUS group 3. At the same time, the M segments of these two viruses had a nucleotide identity of 86%, while the M segment of “isolate 989” has an average nucleotide identity of 87% with the M segment of “isolate Klyukva,” a representative of RUS group 2. Furthermore, within the data set used, no virus showed greater similarity in the M segments than in the S segments with the corresponding genomic fragments of “isolate 989” From this observation, it can be concluded that the ancestor of “isolate 989” arose by reassortment between the ancestor of “strain K27,” from which it inherited the S segment, and the ancestor of the unknown virus, from which it inherited the M and L segments. The results of the analysis by RDP4 software agree with this conclusion, but the tree topology suggests that this event was possibly the reassortment of the M segment.
Phylogenetic analysis to determine the relationships between different viruses is based on the number of different nucleotides in their genomic sequences. From this point of view, the resolving power of the method increases with the number of substitutions. Therefore, it is assumed that an increase in the length of the sequence will allow a more accurate analysis (Yang, 1998). However, due to the presence of reassortment in the genomes (Klempa, 2018), it is only possible to determine the taxonomic relationship of the different PUUVs for individual segments. At the same time, different PUUV genomic segments are not equally represented in the databases. As of June 30th, 2023, there were 514 S segments, 91 M segments and 65 L segments on Genbank. Moreover, usually, if the M and L segments of a virus are known, the S segments are also available, but not conversely. Therefore, a classification based on the L segment, for instance, determines the taxonomic assignment of roughly eightfold fewer viruses in comparison to an analysis of the S segments. Moreover, the S segment rarely undergoes recombination events, probably because of its shorter length compared to other segments. This makes it a good candidate for a basis of a new potential classification system. However, the distribution of pairwise amino acid and nucleotide distances for the protein-coding fragments of the M and L segments (Supplementary Figure S1) resembles the pattern for the S segment (Figure 1). Therefore, a similar “10% nucleotide difference” criterion could be extrapolated for the classification of PUUVs based on M and L segments.
We proposed here to use the criterion “10% nucleotide difference”, which allows an unambiguous subdivision of PUUV into different taxa at the subspecies level. Since this metric is calculated based on the number of differences in the nucleocapsid gene, we propose to refer to these taxa as genotypes. It should be noted that the introduction of similar quantitative criteria has already been proposed for AAV (Beloukhova et al., 2022) and TBEV (Deviatkin et al., 2020). Interestingly, the distribution of the sequences of different viruses (ssRNA (+), ssRNA (−), ssDNA) in the nucleotide/amino acid space was similar, allowing the possibility of a basis for such a subspecies classification of viruses. The clusters of virus pairs (red and cyan dots in Figure 1B) differ in the ratio of synonymous to non-synonymous substitutions. The virus pairs whose nucleoprotein gene sequences differed by less than 10% nucleotides (cyan dots in Figure 1B) had predominantly synonymous substitutions. For example, #MT580935 and #KX757840.1 differed in 9.85% of nucleotides and 0.46% of amino acids. It is important to note that a few critical amino acid changes can be more significant in the way they change the viral phenotype, which can be a limitation of this method. At the same time, the intergenotypic virus pairs (red dots in Figure 1B) showed an accumulation of amino acid substitutions. Such patterns could be explained - viruses exist as a dynamic network of mutational variants (Lauring et al., 2013). Under conditions that are relatively stable for the pathogen (e.g., when the virus is well adapted to the host organism), the viral protein sequence will sooner or later reach its locally optimal state. At the same time, the RNA-dependent RNA polymerase of the virus often makes mistakes when replicating the pathogen genome (Ramsden et al., 2008). As a result, PUUVs do not exist as a homogeneous, discrete entity, but are a collection of different genovariants that differ only minimally from the consensus genome sequence. When polymerase errors lead to non-synonymous substitutions, the mutant viruses move away from the local fitness peak and leave no progeny in competition with viruses closer to the optimal state. At the same time, synonymous substitutions are possible with a number of caveats - for example, if the secondary structure of the RNA is compromised by synonymous substitutions, the fitness of the virus is reduced. However, the environment to which viruses inevitably adapt can change over time, for example, if the host or its way of life changes. In this case, the once optimal virus sequences are no longer optimal. A different viral variant might be better adapted to the new environment and a new protein with slightly different properties might be beneficial (Simmonds et al., 2018). These changes are gradual because the substitutions occur over a long period of time. This means that there are transitional forms of the virus that are closely related both to the group from which they originate and to their progeny group - the cyan dots to the right of the threshold in Figure 1B indicate intragenotypic virus pairs that differ in more than 10% of the nucleotides. An example of such a transitional form is the sequence #JN657228 from the LAT lineage. As mentioned above, the nucleotide difference between sequences #KX815394 and #JN657228 is less than 10%, but the distances between #JN657228 and the other sequences of LAT are more than 10%.
Originally, the nomenclature of Puumala orthohantavirus (PUUV) lineages was predicated upon the geographic locale in which the inaugural representative of the lineage was identified (Sironen et al., 2001; Razzauti et al., 2012; Rosenfeld et al., 2017). In most instances, the geographic distribution of lineages cohered with the nominal region. However, advancements in sequencing technologies have brought to light discrepancies within this nomenclatural framework. Notably, representatives of the FIN lineage were detected in Siberian territory, Russia, whereas those of the RUS lineage were identified in Poland and Latvia, and the LAT lineage was found in Lithuania (Table 1). Thus, the names of the lineages do not always correspond to their actual geographical distribution. The geographical distribution of the different lineages is not a measure of order. The CE, RUS, and FIN lineages are the most expansively distributed geographically, encompassing potential ranges on the order of one million square kilometres. Within such vast expanses, geographically isolated vole populations are inevitably encountered. Conversely, PUUVs affiliated with the DAN lineage, for example, are exclusive to Denmark, whose territory is comparatively smaller. Consequently, the likelihood of encountering an isolated population of hosts for DAN PUUVs is markedly lower than that for CE PUUVs.
Puumala virus evolves along with its hosts - bank voles (Castel et al., 2019). Over time, stochastic mutations accumulate in viral genomes. Those viral strains best adapted to the local ecological parameters of their habitat outcompete alternative variants and subsequently spread over the entire area inhabited by vole populations harbouring the pathogen. When vole populations are separated from each other, each PUUV variant may become increasingly distinct from other virus variants. Such separation is possible due to geographical boundaries - rivers, mountains or adaptation to different ecological conditions. According to the criterion of “10% nucleotide difference”, PUUVs are divided into 21 genotypes, most of which are geographically distinct (Table 1). The exceptions are north-eastern France, Belgium, the Netherlands and southern Germany, where several genotypes belonging to the central European lineage circulate together. This could be explained by the separate occurrence of CE groups 1, 2, 3, 5, 6, 7, and 8 in the past with greater distribution in the present.
Conclusion
In this study, an analysis of all Puumala orthohantaviruses (PUUVs) was performed using nucleotide and protein distance metrics. Viruses with nucleocapsid genes showing less than 10% differences in nucleotide sequence were assigned to the same genotype. Using this established threshold as a delineation criterion for genotypic differentiation, the PUUV species were divided into 21 distinct genotypes. In particular, the CE lineage was subdivided into nine new subgroups, while the RUS lineage was divided into four, the FIN lineage into two and the N-SCA lineage into two groups, each consistent with this defined cut-off.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
AD and PV were responsible for conceptualization. PV secured funding and provided resources for the project. AD supervised the work and handled project administration. AD, MD, and IV were involved in methodology, investigation, software, visualization, and validation. MD and IV were in charge of data curation and formal analysis. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Russian Science Foundation (Grant No 23-64-00002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2024.13168/full#supplementary-material
SUPPLEMENTARY FIGURE S1Scatter plot of amino acid and nucleotide distances for pairs of PUUVs M and L segments.
SUPPLEMENTARY FIGURE S2Similarity plot for the set of sequences with segments separated by 700 N’s. Each point is the percentage of identity compared to the concatenated genomic sequences of “isolate 989” (#OL343606, OL343584, OL343562) within the 900 bp wide sliding window with the 20 bp step.
Footnotes
1.^ https://v-julia.shinyapps.io/recdplot_app/.
2.^ https://github.com/MarselBrukman/PUUV/blob/main/aligned_nucleo_final.fasta.
3.^The alignment file is available at https://github.com/MarselBrukman/PUUV/blob/main/genes_PUUV_separated.fasta.
4.^ https://github.com/MarselBrukman/PUUV/blob/main/genes_PUUV.fasta.
5.^Phylogenetic trees are available at https://github.com/MarselBrukman/PUUV/tree/main/M%20segment%205%20sequences.
6.^Phylogenetic trees are available at https://github.com/MarselBrukman/PUUV/tree/main/M%20segment.
7.^rdp session files are available at https://github.com/MarselBrukman/PUUV/tree/main/rdp4.
8.^Phylogenetic trees in Newick and JPEG formats are available at https://github.com/MarselBrukman/PUUV/tree/main/trees. It should be noted that phylogenetic trees in Newick format contain information about the bootstrap support of the nodes in the trees. This information has been omitted from the figures to simplify visualization.
References
1
Beloukhova M. I. Lukashev A. N. Volchkov P. Y. Zamyatnin A. A. Jr. Deviatkin A. A. (2022). Robust AAV genotyping based on genetic distances in rep gene that are maintained by ubiquitous recombination. Viruses14 (5), 1038. 10.3390/v14051038
2
Blinova E. Deviatkin A. Kurashova S. Balovneva M. Volgina I. Valdokhina A. et al (2022). A fatal case of haemorrhagic fever with renal syndrome in Kursk Region, Russia, caused by a novel Puumala virus clade. Infect. Genet. Evol.102, 105295. 10.1016/j.meegid.2022.105295
3
Blinova E. Deviatkin A. Makenov M. Popova Y. Dzagurova T. (2023). Evolutionary Formation and distribution of puumala virus genome variants, Russia. Emerg. Infect. Dis.29 (7), 1420–1424. 10.3201/eid2907.221731
4
Castel G. Chevenet F. Razzauti M. Murri S. Marianneau P. Cosson J. F. et al (2019). Phylogeography of puumala orthohantavirus in Europe. Viruses11 (8), 679. 10.3390/v11080679
5
Cifuentes-Muñoz N. Salazar-Quiroz N. Tischler N. (2014). Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly. Viruses6 (4), 1801–1822. 10.3390/v6041801
6
Davidyuk Y. Shamsutdinov A. Kabwe E. Ismagilova R. Martynova E. Belyaev A. et al (2020). Prevalence of the Puumala orthohantavirus strains in the pre-kama area of the Republic of Tatarstan, Russia. Pathogens9 (7), 540. 10.3390/pathogens9070540
7
Dekonenko A. Yakimenko V. Ivanov A. Morozov V. Nikitin P. Khasanova S. et al (2003). Genetic similarity of Puumala viruses found in Finland and western Siberia and of the mitochondrial DNA of their rodent hosts suggests a common evolutionary origin. Infect. Genet. Evol.3 (4), 245–257. 10.1016/S1567-1348(03)00088-1
8
Deviatkin A. A. Karganova G. G. Vakulenko Y. A. Lukashev A. N. (2020). TBEV subtyping in terms of genetic distance. Viruses12 (11), 1240. 10.3390/v12111240
9
Fu L. Niu B. Zhu Z. Wu S. Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28 (23), 3150–3152. 10.1093/bioinformatics/bts565
10
Huelsenbeck J. P. Ronquist F. (2001). Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics17 (8), 754–755. 10.1093/bioinformatics/17.8.754
11
Jääskeläinen K. M. Kaukinen P. Minskaya E. S. Plyusnina A. Vapalahti O. Elliott R. M. et al (2007). Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virology79 (10), 1527–1536. 10.1002/jmv.20948
12
Kim J.-A. Kim W. k. No J. S. Lee S. H. Lee S. Y. Kim J. H. et al (2016). Genetic diversity and reassortment of hantaan virus tripartite RNA genomes in nature, the Republic of Korea. PLOS Neglected Trop. Dis.10 (6), e0004650. 10.1371/journal.pntd.0004650
13
Klempa B. (2018). Reassortment events in the evolution of hantaviruses. Virus Genes54 (5), 638–646. 10.1007/s11262-018-1590-z
14
Klempa B. Schmidt H. A. Ulrich R. Kaluz S. Labuda M. Meisel H. et al (2003). Genetic interaction between distinct Dobrava hantavirus subtypes in Apodemus agrarius and A. flavicollis in nature. J. Virology77 (1), 804–809. 10.1128/JVI.77.1.804-809.2003
15
Kumar S. Stecher G. Li M. Knyaz C. Tamura K. (2018). Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35 (6), 1547–1549. 10.1093/molbev/msy096
16
Lauring A. S. Frydman J. Andino R. (2013). The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol.11 (5), 327–336. 10.1038/nrmicro3003
17
Lee S.-H. Kim W. K. No J. S. Kim J. A. Kim J. I. Gu S. H. et al (2017). Dynamic circulation and genetic exchange of a shrew-borne hantavirus, imjin virus, in the Republic of Korea. Sci. Rep.7 (1), 44369. 10.1038/srep44369
18
Lole K. S. Bollinger R. C. Paranjape R. S. Gadkari D. Kulkarni S. S. Novak N. G. et al (1999). Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virology73 (1), 152–160. 10.1128/JVI.73.1.152-160.1999
19
Martin D. P. Murrell B. Golden M. Khoosal A. Muhire B. (2015). RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol.1 (1), 1–5. 10.1093/ve/vev003
20
Nagarajan N. Kingsford C. (2011). GiRaF: Robust, computational identification of influenza reassortments via graph mining. Nucleic Acids Res.39 (6), e34. 10.1093/nar/gkq1232
21
Nguyen L.-T. Schmidt H. A. von Haeseler A. Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32 (1), 268–274. 10.1093/molbev/msu300
22
Pérez-Losada M. Arenas M. Galán J. C. Palero F. González-Candelas F. (2015). Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol.30, 296–307. 10.1016/j.meegid.2014.12.022
23
Plyusnin A. Vapalahti O. Vaheri A. (1996). Hantaviruses: Genome structure, expression and evolution. J. General Virology77 (11), 2677–2687. 10.1099/0022-1317-77-11-2677
24
Rambaut A. (2020). FigTree 1.4.4.
25
Ramsden C. Melo F. L. Figueiredo L. M. Holmes E. C. Zanotto P. M. (2008). High rates of molecular evolution in hantaviruses. Mol. Biol. Evol.25 (7), 1488–1492. 10.1093/molbev/msn093
26
Razzauti M. Plyusnina A. Henttonen H. Plyusnin A. (2008). Accumulation of point mutations and reassortment of genomic RNA segments are involved in the microevolution of Puumala hantavirus in a bank vole (Myodes glareolus) population. J. General Virology89 (7), 1649–1660. 10.1099/vir.0.2008/001248-0
27
Razzauti M. Plyusnina A. Niemimaa J. Henttonen H. Plyusnin A. (2012). Co-Circulation of two puumala hantavirus lineages in Latvia: a Russian lineage described previously and a novel Latvian lineage. J. Med. Virology84 (2), 314–318. 10.1002/jmv.22263
28
Razzauti M. Plyusnina A. Sironen T. Henttonen H. Plyusnin A. (2009). Analysis of puumala hantavirus in a bank vole population in northern Finland: Evidence for co-circulation of two genetic lineages and frequent reassortment between strains. J. General Virology90 (8), 1923–1931. 10.1099/vir.0.011304-0
29
Rosenfeld U. M. Drewes S. Ali H. S. Sadowska E. T. Mikowska M. Heckel G. et al (2017). A highly divergent Puumala virus lineage in southern Poland. Archives Virology162 (5), 1177–1185. 10.1007/s00705-016-3200-5
30
Sibold C. Meisel H. Krüger D. H. Labuda M. Lysy J. Kozuch O. et al (1999). Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J. Virology73 (1), 667–675. 10.1128/JVI.73.1.667-675.1999
31
Simmonds P. Aiewsakun P. Katzourakis A. (2018). Prisoners of war — host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. Prepr.17, 321–328. 10.1038/s41579-018-0120-2
32
Sinisalo M. Vapalahti O. Ekblom-Kullberg S. Laine O. Mäkelä S. Rintala H. et al (2010). Headache and low platelets in a patient with acute leukemia. J. Clin. Virology48 (3), 159–161. 10.1016/j.jcv.2010.02.015
33
Sironen T. Vaheri A. Plyusnin A. (2001). Molecular evolution of puumala hantavirus. J. Virology75 (23), 11803–11810. 10.1128/JVI.75.23.11803-11810.2001
34
Szabó R. Radosa L. Ličková M. Sláviková M. Heroldová M. Stanko M. et al (2017). Phylogenetic analysis of Puumala virus strains from Central Europe highlights the need for a full-genome perspective on hantavirus evolution. Virus Genes53 (6), 913–917. 10.1007/s11262-017-1484-5
35
Vaheri A. Strandin T. Hepojoki J. Sironen T. Henttonen H. Mäkelä S. et al (2013). Uncovering the mysteries of hantavirus infections. Nat. Rev. Microbiol.11 (8), 539–550. 10.1038/nrmicro3066
36
Vakulenko Y. Deviatkin A. Drexler J. F. Lukashev A. (2021). Modular evolution of coronavirus genomes. Viruses13 (7), 1270. 10.3390/v13071270
37
Wickham H. (2016). ggplot2. Cham: Springer International Publishing. (Use R!). 10.1007/978-3-319-24277-4
38
Yang Z. (1998). On the best evolutionary rate for phylogenetic analysis. Syst. Biol.47 (1), 125–133. 10.1080/106351598261067
Summary
Keywords
genotype, PUUV, puumala virus, hantavirus, taxonomy
Citation
Dashian MA, Volkhin IA, Volchkov PY and Deviatkin AA (2024) Genotypic differentiation and evolutionary dynamics of puumala orthohantavirus (PUUV). Acta Virol. 68:13168. doi: 10.3389/av.2024.13168
Received
23 April 2024
Accepted
11 September 2024
Published
20 September 2024
Volume
68 - 2024
Edited by
Boris Klempa, Slovak Academy of Sciences, Slovakia
Reviewed by
Yoonsoo Hahn, Chung-Ang University, Republic of Korea
Won-Keun Kim, Hallym University, Republic of Korea
Updates
Copyright
© 2024 Dashian, Volkhin, Volchkov and Deviatkin.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilya A. Volkhin, ilyavolkhin2@gmail.com; Andrei A. Deviatkin, devyatkin_aa@academpharm.ru
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.