- School of Science, Engineering and Environment, University of Salford, Salford, United Kingdom
A novel morbillivirus and a novel betaherpesvirus are reported in the Wood Mouse (Apodemus sylvaticus) in the western United Kingdom (UK). The two viruses were found coinfecting an underweight host with abnormalities of the liver and were detected via deep sequencing of lung RNA and de novo assembly of substantial genome fragments. The phylogenetic affinities of the novel viruses are characterised based on their relationships to existing database sequences.
Introduction
The virome of wild rodents in the UK, and globally, is of intrinsic ecological importance and also of great significance as a reservoir from which zoonotic and trans-species infectious diseases may emerge (Wu et al., 2018). The viral burden of UK wildlife is only partially documented but a more complete picture would be of value in assessing public and animal health risks (Carlson et al., 2021) and in understanding the regulation and health of wild populations (Lochmiller, 1996). In this report two novel viruses are documented from a wild rodent, the Wood Mouse (Apodemus sylvaticus), in the UK. These viruses were discovered via de novo assembly of substantial genomic fragments, working from Illumina short-read sequences of host lung RNA. The context of the discovery indicates the likelihood of substantial undiscovered diversity.
Method
A single mature male A. sylvaticus was captured in a forestry plantation near the confluence of the Afon Tarennig and the Afon Gwy/River Wye (52.426773, −3.707703) in July 2011 by humane live-trapping and killed via a UK Home Office approved method. In the UK, Apodemus mice are represented only by A. sylvaticus and A. flavicollis (see Mathews et al., 2018). The latter is not known to occur in the study area, although it can occur in adjacent areas of Mid Wales (Mathews et al., 2018). To avoid any possibility of misidentification, the present specimen was initially positively identified on the basis of morphological criteria, primarily ventral colouration and foot length (Flowerdew, 1984). This identification was consistent with de novo assembled host transcripts derived from the RNAseq data that are described further below. For example, 840 base pair fragments of coding sequences for A. flavicollis toll-like receptor 5 (tlr5) haplotypes in National Center for Biotechnology Information (NCBI) GenBank (Sayers et al., 2022) (GenBank accession numbers: OM365774-6) share 98.2%–98.3% identity with a corresponding transcript fragment for the present specimen. In comparison, this same fragment from the present specimen shares 99.5% identity with a homologous fragment predicted from the A. sylvaticus NCBI (Sayers et al., 2022) RefSeq genome (XM_052201127). The captured specimen was underweight, with a negative residual of c.2.5 g from a weight on length regression of mature males (n = 12) measured at nearby sites in summer 2011. Furthermore, upon internal examination, there were multiple adhesions of the liver to the abdominal cavity wall and abnormal, fibrotic growth of liver tissue investing the anteroventral surface of the right kidney. No other abnormalities were observed. Fresh, apparently healthy, lung tissue was removed and stored in RNA stabilization solution. Following total RNA extraction from this material and poly (A)-focussed sequencing library construction (originally intended as a control for a separate study, and not aimed primarily at virus discovery in the present specimen), the library was sequenced on an Illumina Novaseq machine (at Azenta Life Sciences, UK) yielding c. 171 million 150 base pair paired end reads. The reads were mapped to a Mus musculus genome (mm9) using BBMap (Bushnell et al., 2017) and unmapped reads were de novo assembled employing rnaviralSPAdes (Meleshko and Korobeynikov, 2023). Assembled contigs were then searched against the NCBI (Sayers et al., 2022) RefSeq viral genomes database via discontiguous megablast (Camacho et al., 2009) at a 40% identity cut-off, with only the strongest hit per contig considered. Substantial alignments (>200 base pairs) to viral sequences were further investigated via individual blastn (Camacho et al., 2009) searches against the NCBI (Sayers et al., 2022) nt database and only unambiguous matches to viral sequences (excluding phages and retroviruses) were considered further.
Results and discussion
We found clear evidence for the presence of two novel viruses in the lungs of the Wood Mouse from our study site. In one case, two large genome fragments were assembled of 1684 (GenBank accession number: PP188092) and 1592 base pairs (PP188093). These unambiguously clustered with rodent-infecting members of the Morbillivirus genus in phylogenetic analyses (see Figure 1). Classical morbilliviruses occur in larger mammals and are well-known agents of serious diseases, such as measles, canine distemper, phocine distemper, rinderpest, peste des petits ruminants and cetacean morbillivirus (Seki and Takeda, 2022; Libbey and Fujinami, 2023). The present discovery adds to the divergent members of the Morbillivirus genus that have recently been described in rodents (Debat, 2022; Vanmechelen et al., 2022; Chen et al., 2023) and bats (Wells et al., 2022; Ikegame et al., 2023). The novel morbillivirus (Tarennig A. sylvaticus morbillivirus) is closest to, and might be considered a well differentiated strain of, Gierle Apodemus virus (Vanmechelen et al., 2022). The 1684 base pair assembled fragment encompassed a full 1572 base pair open reading frame coding for a 524 amino acid nucleocapsid-like protein with 95% residue similarity to the 526 amino acid nucelocapsid protein (UQM99541.1) in Gierle Apodemus Virus. The 1592 base pair fragment encompassed a full 1488 base pair open reading frame coding for a 496 amino acid phosphoprotein-like protein with 89% residue similarity to the 496 amino acid phosphoprotein in Gierle Apodemus Virus (UQM99543.1).
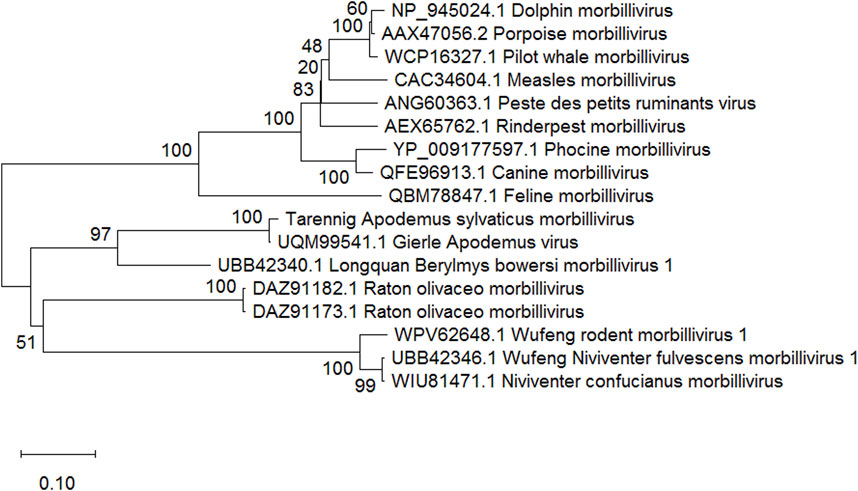
Figure 1. Neighbour joining tree (Saitou and Nei, 1987) of representative morbillivirus nucleocapsid proteins, based on 388 amino acid positions aligned using ClustalW (Thompson et al., 1994). Percentage bootstrap values supporting clusters are shown next to corresponding branches. GenBank accession numbers and a taxon name are shown for each sequence. The scale bar shows amino acid substitutions per site. Phylogenetic analysis was conducted in MEGA Version 10.0.05 (Kumar et al., 2018).
In another case, two large genome fragments of 834 base pairs (PP188094) and 546 base pairs (PP188095) were assembled that clustered with murine betaherpesviruses (Muromegalovirus (Walker et al., 2022) or Cytomegalovirus) in phylogenetic analyses (see Figure 2). The novel virus (Tarennig A. sylvaticus betaherpesvirus) was close to, but well differentiated from, Murid betaherpesvirus 1. The larger genome fragment contained an 816 bp part of an open reading frame, corresponding to a 272 amino acid sequence, with greatest similarity to the m142 protein of Murid betaherpesvirus 1 (AWV68063.1; 67% amino acid identity across a 237 aa aligned region). The smaller genome fragment contained a 543 base pair part of an open reading frame, corresponding to a 181 amino acid sequence, with greatest similarity to the m85 protein of Murid betaherpesvirus 1 and its equivalent (capsid triplex subunit 2) in Mastomys natalensis cytomegalovirus 3 (WEG69771.1; with 85% identity over a 158 amino acid aligned region in this case). As far as can be determined this is the first record of a betaherpesvirus in the Wood Mouse in the U.K. or elsewhere, although there is a disputed claim (Kim et al., 1974) that a murine cytomegalovirus derived from Wood Mice was adapted to replicate in human cells in vitro in the 1960s (Raynaud et al., 1969). Numerous other betaherpesviruses (Ehlers et al., 2007; Tarlinton et al., 2011; Ntumvi et al., 2018) have been reported in rodents on the basis of small sequence fragments of genes not recovered here, including in A.flavicollis (Yellow-necked Mice) in Germany (Ehlers et al., 2007). It was secondarily possible to recover two reads corresponding to the betaherpesvirus DNA polymerase gene, which has been targeted in some viral discovery studies, and these were most similar to A. flavicollis cytomegalovirus 2 (EF125063.1; 86%–87% nucleotide identity) and then to Murid betaherpes virus 1 isolates (78%–81% nucleotide identity).

Figure 2. Neighbour joining tree (Saitou and Nei, 1987) of representative betaherpesvirus m142 and related proteins, based on 194 amino acid positions aligned using ClustalW (Thompson et al., 1994). Percentage bootstrap values supporting clusters are shown next to corresponding branches. GenBank accession numbers and a protein identifier/taxon name are shown for each sequence. The scale bar shows amino acid substitutions per site. Phylogenetic analysis was conducted in MEGA Version 10.0.05 (Kumar et al., 2018).
It is recognized that the present study is based on a limited sample (a single tissue from a single host) and that further studies are required to reveal the epidemiological characteristics and biology of the discovered viruses. Nonetheless, the current records usefully extend our knowledge of the virome in UK wild mammals. It is also noted that neither virus was straightforwardly detected via the analysis of short reads with taxonomic classifier softwares under a range of conditions and that such classifiers were prone to false positives due to host-derived rRNA hits. The ready detection of new viruses via the methods employed, even from a single arbitrarily sampled host specimen, and the high level of nucleotide divergence of the viruses from their closest relatives in databases, are suggestive of considerable undiscovered viral diversity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) are as follows: https://www.ncbi.nlm.nih.gov/genbank/, PP188092 https://www.ncbi.nlm.nih.gov/genbank/, PP188093 https://www.ncbi.nlm.nih.gov/genbank/, PP188094 https://www.ncbi.nlm.nih.gov/genbank/, PP188095.
Ethics statement
The study complied with the ethics review procedure at Aberystwyth University at the time the work was carried out and was conducted in accordance with the relevant local legislation and institutional requirements.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Fieldwork was supported by Aberystwyth University and sequencing work was supported by the University of Salford.
Acknowledgments
Grateful acknowledgement is made to the UK Forestry commission, for permission to trap at the field site in 2011, and to the Galaxy Europe server, on which some bioinformatic analyses were carried out.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bushnell, B., Rood, J., and Singer, E. (2017). BBMerge – accurate paired shotgun read merging via overlap. Plos one 12, e0185056. doi:10.1371/journal.pone.0185056
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10, 421. doi:10.1186/1471-2105-10-421
Carlson, C. J., Farrell, M. J., Grange, Z., Han, B. A., Mollentze, N., Phelan, A. L., et al. (2021). The future of zoonotic risk prediction. Philosophical Trans. R. Soc. Lond. Ser. B, Biol. Sci. 376, 20200358. doi:10.1098/rstb.2020.0358
Chen, Y.-M., Hu, S.-J., Lin, X.-D., Tian, J.-H., Lv, J.-X., Wang, M.-R., et al. (2023). Host traits shape virome composition and virus transmission in wild small mammals. Cell 186, 4662–4675.e12. doi:10.1016/j.cell.2023.08.029
Debat, H. J. (2022). A South American mouse morbillivirus provides insight into a clade of rodent-borne morbilliviruses. Viruses 14, 2403. doi:10.3390/v14112403
Ehlers, B., Küchler, J., Yasmum, N., Dural, G., Voigt, S., Schmidt-Chanasit, J., et al. (2007). Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J. Virol. 81, 8091–8100. doi:10.1128/jvi.00255-07
Flowerdew, J. (1984). Woodmice and yellow-necked mice (mammal society series). Oswestry, UK: Nelson.
Ikegame, S., Carmichael, J. C., Wells, H., Furler O’Brien, R. L., Acklin, J. A., Chiu, H. P., et al. (2023). Metagenomics-enabled reverse-genetics assembly and characterization of myotis bat morbillivirus. Nat. Microbiol. 8, 1108–1122. doi:10.1038/s41564-023-01380-4
Kim, K. S., Sapienza, V., and Carp, R. I. (1974). Comparative studies of the Smith and Raynaud strains of murine cytomegalovirus. Infect. Immun. 10, 672–674. doi:10.1128/iai.10.3.672-674.1974
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Libbey, J. E., and Fujinami, R. S. (2023). Morbillivirus: A highly adaptable viral genus. Heliyon 9, e18095. doi:10.1016/j.heliyon.2023.e18095
Lochmiller, R. L. (1996). Immunocompetence and animal population regulation. Oikos 76, 594–602. doi:10.2307/3546356
Mathews, F., Kubasiewicz, L. M., Gurnell, J., Harrower, C. A., McDonald, R. A., and Shore, R. F. (2018). A review of the population and conservation status of British mammals. Peterborough, UK: Natural England.
Meleshko, D., and Korobeynikov, A. (2023). Benchmarking state-of-the-art approaches for norovirus genome assembly in metagenome sample. Biology 12, 1066. doi:10.3390/biology12081066
Ntumvi, N. F., Mbala Kingebeni, P., Tamoufe, U., Kumakamba, C., Ndze, V., Ngay Lukusa, I., et al. (2018). High herpesvirus diversity in wild rodent and shrew species in Central Africa. Intervirology 61, 155–165. doi:10.1159/000493796
Raynaud, J., Atanasiu, P., Barreau, C., and Jahkola, M. (1969). Adaptation d’un virus cytomégalique provenant du Mulot (Apodemus sylvaticus) sur différentes cellules hétérologues, y compris les cellules humaines. Comptes Rendus Hebd. Séances L’Académie Sci.269, 104–106.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi:10.1093/oxfordjournals.molbev.a040454
Sayers, E. W., Bolton, E. E., Brister, J. R., Canese, K., Chan, J., Comeau, D. C., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50 (D1), D20–D26. doi:10.1093/nar/gkab1112
Seki, F., and Takeda, M. (2022). Novel and classical morbilliviruses: current knowledge of three divergent morbillivirus groups. Microbiol. Immunol. 66, 552–563. doi:10.1111/1348-0421.13030
Tarlinton, R. E., Tate, H. F. J., Mohallal, E. M. E., Daly, J. M., and Behnke, J. M. (2011). Betaherpesvirus sequences in eastern spiny mice and wagner’s dipodils. J. Wildl. Dis. 47, 958–962. doi:10.7589/0090-3558-47.4.958
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi:10.1093/nar/22.22.4673
Vanmechelen, B., Meurs, S., Horemans, M., Loosen, A., Joly Maes, T., Laenen, L., et al. (2022). The characterization of multiple novel paramyxoviruses highlights the diverse nature of the subfamily Orthoparamyxovirinae. Virus Evol. 8, eac061. doi:10.1093/ve/veac061
Walker, P. J., Siddell, S. G., Lefkowitz, E. J., Mushegian, A. R., Adriaenssens, E. M., Alfenas-Zerbini, P., et al. (2022). Recent changes to virus taxonomy ratified by the international committee on taxonomy of viruses (2022). Arch. Virol. 167, 2429–2440. doi:10.1007/s00705-022-05516-5
Wells, H. L., Loh, E., Nava, A., Solorio, M. R., Lee, M. H., Lee, J., et al. (2022). Classification of new morbillivirus and jeilongvirus sequences from bats sampled in Brazil and Malaysia. Arch. Virol. 167, 1977–1987. doi:10.1007/s00705-022-05500-z
Keywords: Apodemus sylvaticus, betaherpesvirus, liver disease, morbillivirus, pulmonary
Citation: Jackson JA (2024) A novel morbillivirus and a novel betaherpesvirus infecting the Wood Mouse in the UK. Acta Virol. 68:13177. doi: 10.3389/av.2024.13177
Received: 24 April 2024; Accepted: 26 July 2024;
Published: 08 August 2024.
Edited by:
Boris Klempa, Slovak Academy of Sciences, SlovakiaReviewed by:
Vanessa Herder, MRC-University of Glasgow Centre For Virus Research (MRC), United KingdomĽuboš Korytár, University of Veterinary Medicine and Pharmacy in Košice, Slovakia
Copyright © 2024 Jackson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph A. Jackson, ai5hLmphY2tzb25Ac2FsZm9yZC5hYy51aw==
 Joseph A. Jackson
Joseph A. Jackson