Abstract
The non-uniform usage of synonymous codons occurs in genomes of all organisms, including DNA and RNA viruses. The preferential selection of a codon at the expense of other synonymous codons within the same group is known as Codon Usage Bias. The understanding of this bias assists in unveiling the factors driving molecular evolution, as defined by the selection-mutation-drift theory. According to this model, molecular evolution is predominantly driven by mutation, natural selection, and genetic drift. Nevertheless, elements like nucleotide composition, gene length, and protein secondary structure also contribute to this process. Comprehensive genomic analyses that highlight the codon usage preference of the African Swine Fever Virus (ASFV) are infrequent. ASFV, a hemorrhagic and highly contagious viral disease, almost invariably results in 100% fatality among infected pigs and wild boars. This study, therefore, embarked on a thorough examination of codon usage patterns in ASFV’s complete genomic sequences, an endeavor of great relevance to molecular evolution studies, complex transmission models, and vaccine research. For an exhaustive evaluation of ASFV’s whole-genome codon usage, we used parameters like ENC, RSCU, and CAI. A Principal Component Analysis was carried out to reaffirm the interconnected RSCU lineages based on the continent, and their evolutionary relationships were later elucidated through phylogenetic tree construction. ASFV emerged as a low-biased codon user (ENC = 52.8) that is moderately adapted to its host. Its genome has a high AT composition (64.05%), suggesting the impact of mutational pressure on genomic evolution. However, neutrality plot analysis revealed natural selection’s slight supremacy over mutational pressure. The low codon bias (>45) implies ASFV’s diverse usage of synonymous codons within a given codon family, allowing for effective translation and subsequent successful viral replication cycles. Its moderate adaptation (CAI = 0.56) permits the virus to infect a range of hosts, including reservoirs such as warthogs and bush pigs. To the best of our knowledge, this is the pioneering report providing a comprehensive examination of ASFV’s complete genomic sequences. Consequently, research focusing on viral gene expression and regulation, gene function prediction, parasite-host interaction, immune dysfunction, and drug and vaccine design may find this report to be a valuable resource.
Introduction
African Swine Fever (ASF) constitutes a virulent, infectious hemorrhagic fever afflicting both domestic pigs and wild boars. This deadly disease originates from the African Swine Fever Virus (ASFV), the solitary DNA arbovirus and exclusive member of the Asfivirus genus, nested within the Asfarviridae family (Dixon et al., 2012; Jia et al., 2017; Teklue et al., 2020). Depending on the ASFV strain, morbidity and mortality rates may escalate to 100%, making ASF a severe obstacle for global pig farming and, consequently, worldwide food and nutrition security (Zhang et al., 2022). ASFV is a double-stranded DNA virus ranging from 170 to 193 kbp in length and encompassing between 151 and 167 open reading frames depending on the strain. The virus presents a complex, multi-layered icosahedral virion structure (Jia et al., 2017; Wang et al., 2021; Zheng et al., 2022). It shares characteristics with other large nucleocytoplasmic DNA viruses such as poxvirus and iridovirus (Iyer et al., 2001; Loh et al., 2009).
Through nucleotide sequencing of the B646L gene, which encodes the p72 major capsid protein, twenty-four (I to XXIV) ASFV genotypes have been identified, all found in Africa. ASF remains enzootic in sub-Saharan Africa and Sardinia, Italy (Sang et al., 2020). However, it has migrated beyond these traditional confines, infiltrating regions such as Caucasia, the European Union, Asia, Oceania, and South America, and has recently resurfaced in North America’s Haiti and the Dominican Republic (Alkhamis et al., 2018; Ata et al., 2022; Ayanwale et al., 2022; Chen et al., 2022). In 2018, ASF penetrated China, the world’s leading pork producer, decimating extensive pig populations and jeopardizing global food and protein security. Since there are no treatments or vaccines for ASF, the primary means of containment are quarantine, culling, and rigorous movement restrictions (Salguero, 2020; Urbano and Ferreira, 2020).
Codon Usage Bias (CUB) refers to the phenomenon wherein synonymous codons within a codon family are employed at diverse frequencies. Synonymous codons are substitute codons in the same group that offer a different coding sequence for a certain amino acid (Zhang et al., 2011; Deb et al., 2021a). The genetic code’s degeneracy facilitates this, meaning that a single amino acid can be coded by multiple codons within the same family (Nasrullah et al., 2015; Tian et al., 2018). This redundancy in the genetic code is crucial in modulating the efficacy and precision of protein synthesis while preserving the amino acid sequence of the protein. Apart from methionine and tryptophan, this degeneracy enables the remaining 18 amino acids to be encoded by 61 triplet codons (Nasrullah et al., 2015; Yao et al., 2020).
Numerous DNA and RNA viruses exhibit CUB, with some, such as the Hepatitis A Virus, showing significant bias (ENC = 39.78) (Andrea et al., 2011; Bera et al., 2017). In contrast, other viruses, such as PDCoV (ENC = 52.69) (Peng et al., 2022), SARS-CoV-2 (ENC = 45.38) (Hou, 2020), PEDV (ENC = 48.04) (Yu et al., 2021a), CHIKV (ENC = 55.56) (Butt et al., 2014), ZIKV (ENC = 53.32) (Wang et al., 2016), Classical Swine Fever Virus (ENC = 51.7) (Tao et al., 2009), H7N9 Influenza A Virus (ENC = 51) (Sun et al., 2020), and Bluetongue (ENC = 57.9) (Yao et al., 2020), display weak codon usage bias.
Several factors can potentially influence codon usage patterns, including mutational pressure, natural selection, genetic drift, gene length, nucleotide and amino acid composition, secondary protein structure, replication, and transcription factors, the hydrophobicity and hydrophilicity of the protein, and environmental conditions (Butt et al., 2014; Zhang et al., 2018). However, under the selection-mutation-genetic drift model, natural selection, mutational pressure, and genetic drift primarily account for variations in codon usage among diverse organisms (Deb et al., 2021a). For many RNA and DNA viruses, including Foot-and-Mouth Virus, Herpes Virus, and Rubella Virus, mutational bias is more pronounced than natural selection. Conversely, natural selection is instrumental in the evolution of certain viruses, such as those in the Parvoviridae family, Zika, Henipa, and Influenza A viruses. Therefore, despite mutational pressure’s ongoing significance as an evolutionary force, it is not the sole influencer (Shi et al., 2013; Buttv et al., 2014; Sun et al., 2020).
Viral genomes diverge from their prokaryotic and eukaryotic counterparts due to their dependence on host cell machinery for genome replication and protein synthesis. This co-evolution impacts viral gene expression, protein synthesis, pathogenicity, and immune evasion (Butt et al., 2014; Kumar et al., 2021). Hence, a comprehensive exploration of CUB is necessary for understanding viral evolution, adaptation, and genomic features. CUB patterns serve as key indicators of genomic evolution (Chen et al., 2014; He et al., 2019). Consequently, this study examined the CUB of ASFV, with the findings expected to facilitate understanding of viral strain evolutionary trends, elucidate transmission pathways, and determine the speed of evolution. Moreover, such knowledge is crucial for predicting gene function, expression, regulation, and designing vaccines.
Materials and methods
Complete genome sequences of ASFV isolates/strains were retrieved from the GenBank database via the web portal at https://www.ncbi.nlm.nih.gov/data-hub/genome/. For an in-depth understanding of ASFV codon usage bias, only viruses with complete genomic information were considered in this study. Following the rigorous elimination of redundancy and potential recombination, 54 distinct strains (n = 54) from the collected samples (n = 174) were selected and included in this analysis (Supplementary Table S1). Ten Open Reading Frames (ORFs) of each of the 54 retained strains were obtained from the Open Reading Frame Finder available at https://www.ncbi.nlm.nih.gov/orffinder/ for codon usage parameter estimation.
During the selection of sequences, the following criteria were used; sequences with only ATG as a start codon were retrieved, sequences with a minimal length of less than 75 were not retrieved, and sequences with nested ORFs were also not retrieved. For simplicity, the 10 longest ORFs, as one of the options displayed in an ORF Finder, were selected. However, sequences that had no terminating codons were removed and replaced with the ones with stop codons.
Fundamental indices were computed using the online CAIcal server available at http://genomes.urv.es/CAIcal/ (Puigbò et al., 2008). These parameters include Relative Synonymous Codon Usage (RSCU), Effective Number of Codons (ENC), Codon Adaptation Index (CAI), and Relative Codon Deoptimization Index (RCDI). Sus scrofa domestica’s coding sequences with 22094 codons used as a host reference genome, in this regard, were retrieved from the Codon Usage Database available at https://www.kazusa.or.jp/codon/ (Nakamura et al., 2000). The Similarity Index (SiD) and Second Parity Rule Analysis, however, were executed using the vhcub package in R software version 4.1.3, which can be downloaded at https://cran.r-project.org/web/packages/vhcub/ (Anwar et al., 2020). Hydropathicity and aromaticity indices were calculated using CodonW 1.4.2 version available at http://codonw.sourceforge.net/. Principal Component Analysis (PCA), which extracts major trends of variations among the strains by transforming RSCU values into uncorrelated variables, and the correlation analysis between principal components and codon usage indices via Spearman’s rank method were performed using R software. The selected packages for PCA were factoextra and FactoMineR, supported by ggplot2 (Husson et al., 2022; Kassambara and Mundt, 2022; Wickham et al., 2022).
For phylogenetic analysis, sequence data was compiled and edited using BioEdit (version 7.0.9.0). Multiple sequence alignment (MAS) was executed online via MAFFT software available at https://www.ebi.ac.uk/Tools/msa/mafft/, while the phylogenetic tree was constructed using MEGA version 11, downloadable at https://www.megasoftware.net/. The subsequent section provides a detailed account of the codon usage indices and other vital components.
Nucleotide composition
The nucleotide compositional constraints of the entire genome ORFs were assessed using the CAIcal server (http://genomes.urv.es/CAIcal/) (Puigbò et al., 2008). This analysis involved the calculation of various measures including the overall nucleotide frequency, the nucleotide frequency at the third position of synonymous codons, the average frequency of G and C nucleotides at the third codon position (GC3), and the average frequency of G and C nucleotides at the first and second codon positions(GC12). The termination codons TGA, TAG, and TAA, as well as the ATG codon for Methionine and the TGG codon for Tryptophan, were excluded from this analysis.
Relative synonymous codon usage
Relative Synonymous Codon Usage (RSCU) refers to the differential usage of synonymous codons that code for the same amino acid. Essentially, RSCU is the ratio of the observed frequency to the expected frequency of a codon, assuming that all codons for a particular amino acid are used randomly (Sharp and Li, 1986). The RSCU value is unaffected by amino acid composition and is often used to estimate codon usage bias. A higher RSCU value implies a higher frequency of codon usage and a stronger codon usage bias. A codon is said to have a positive codon usage bias if its RSCU value is >1.0, and a negative codon usage bias if its RSCU value is <1.0. Codons with RSCU values <0.6 are considered under-represented, whereas those with values >1.6 are over-represented. Codons with RSCU values of 1.0 are considered unbiased, and used at an equal frequency (Sharp and Li, 1986).
The following formula is used to calculate RSCU values for each strain:where Xij represents the occurrence frequency of the jth codon for the ith amino acid (any Xij with a value of zero is at random assigned a value of 0.5), and ni represents the number of codons for the ith amino acid (ith codon family).
Principal component analysis
Principal component analysis (PCA) is a widely used statistical method for analyzing CUB via a dimension reduction strategy (Sharp and Li, 1986; Wold et al., 1987). This analysis reveals the main trends among the correlated variables (in this case, codons) across different ASFV strains. The RSCU values for each strain are represented by a 59-dimensional vector of codons. PCA then transforms these RSCU values into uncorrelated variables captured by principal components (Todorov et al., 2018). After this, the factors influencing codon usage bias are assessed via correlation analysis between the principal axes (PC1 & PC2) and codon usage parameters.
Effective number of codons
The effective number of codons (ENC) provides a measure of the extent to which codon usage in a gene or genome deviates from equal use of synonymous codons (Wright, 1990). In ENC analysis, each codon is assigned an ENC value. ENC values range from 20 to 61. Unlike RSCU, a higher ENC value indicates less codon usage bias. Neither the amino acid content nor the gene length impacts the effective number of codons. ENC values less than or equal to 45 generally signify strong codon usage bias (Comeron and Aguade, 1998).
The standard ENC values were calculated using the formula:where; F (i = 2, 3, 4, 6) represents the mean Fi values for the i-fold degenerate amino acids. The Fi value represents the likelihood that two randomly chosen codons for an amino acid are identical. Fi is determined as follows:where i is the type number of the synonymous codon family to which the particular amino acid belongs, nj is the number of observed counts of the jth codon for that amino acid, and n is the total number of observed codon counts for the same amino acid.
Codon adaptation index
The Codon Adaptation Index (CAI) is a measure used to quantify the similarity between the codon usage of viral genes or genomes and their hosts (Sharp and Li, 1987). It is a widely used method for assessing codon usage bias due to natural selection. It indicates the adaptation of the virus to the host. The CAI value ranges from 0 to 1, with higher values suggesting stronger host adaptability. The CAI values were computed using the CAIcal server available at http://genomes.urv.es/CAIcal/ to estimate the ASFV codon adaptation to its host. The reference dataset of synonymous codon usage was downloaded from the Codon Usage Database available at https://www.kazusa.or.jp/codon/ (Nakamura et al., 2000).
Relative Codon Deoptimization Index
The Relative Codon Deoptimization Index (RCDI) quantifies the similarity between a gene’s codon usage and the codon usage of a reference genome (Mueller et al., 2006). It can also be employed to assess the speed of viral gene translation within a host genome and estimate the co-evolution of the virus with its host. RCDI values near 1 indicate a virus that is highly adapted to its host and follows the codon usage pattern of its host. RCDI values greater than 1 suggest the virus is less well-adapted to the host or has codon usage patterns different from its host (Butt et al., 2016). The RCDI values were computed using the RCDI/eRCDI server available at http://genomes.urv.es/CAIcal/ (Puigbò et al., 2010). The codon usage pattern for Sus scrofa domesticus was retrieved from the Codon Usage Database for use as reference, as was done for the CAI calculation (Nakamura et al., 2000).
Similarity index analysis
The similarity index [SiD or D (A, B)] was utilized to estimate the impact of overall host codon usage patterns on virus gene expression (Zhou et al., 2013). SiD ranges between 0 and 1, with a higher value indicating a stronger influence of the host on virus codon usage. To further investigate the influence of Sus scrofa codon usage patterns on ASFV codon usage, the similarity index was computed as follows:where R(A, B) is defined as the cosine of the angle between the spatial vectors A and B, representing the degree of similarity between the overall usage patterns of the virus and host codons. ai represents the RSCU value for a specific codon from synonymous codons of the ASFV strain. bi represents the RSCU value for the same codon in the host. D (A, B) represents the potential impact of the host’s overall codon usage on the ASFV strain. This value ranges from 0 to 1.0, with a higher value suggesting that the host has a strong influence on codon usage.
Hydropathicity and aromaticity indices
The GRAVY score for hydropathicity and the Aroma score for aromaticity were employed to estimate the biochemical properties of translated proteins (Lobry and Gautier, 1994). GRAVY stands for General Average Hydropathicity; it ranges from −2 to 2, where negative and positive values respectively indicate hydrophilic and hydrophobic amino acids. The Aroma score indicates the presence of aromatic amino acids such as tryptophan, tyrosine, and phenylalanine in an encoded protein. Hydropathy and aroma values provide insight into the biochemical significance of the synthesized protein, indicating the effects of natural selection on a viral genome and, consequently, on the virus’s codon usage (Chen et al., 2014; Yu et al., 2021b).
Neutral evolution analysis
In ASFV, neutral analysis was employed to assess the changing influences of mutational pressure and natural selection (Sueoka, 1988). This study presents the GC12 values of the synonymous codons plotted against the GC3 values. To analyze the evolutionary characteristics of mutational pressure and natural selection in ASFV, the GC12 or GC3 values are plotted against evolutionary time. The slope of a simple regression line signifies the evolutionary speed of mutational and natural selection pressures. The influence of mutational pressure and natural selection on base composition can be assessed by examining the correlation between the GC content at the first and second codon positions (GC12) and that at the third codon position (GC3). As a result, the online software tool available at http://genomes.urv.es/CAIcal/ was used to determine GC12 and GC3, and regression analysis was then conducted using R. The mutation-selection equilibrium coefficient, representing the evolutionary speed of mutational pressure and natural selection pressure, is the slope (regression coefficient) of the regression line. If all points spread diagonally (slope = 1) and the correlation between the GC3 and GC12 variables is statistically significant, mutation is the primary factor influencing codon usage. Otherwise, if the regression line is parallel or inclined towards the horizontal axis (near-zero slope), the selection is considered the dominant factor (Sueoka, 1988).
ENC-GC3 plot analysis
ENC values were plotted against GC3 values in an ENC-GC3 plot to discern the primary influences on codon usage bias (Wright, 1990). In this diagram, the ENC value forms the y-axis and the GC3 value constitutes the x-axis. Points located on or slightly below the expected curve suggest mutational pressure as the principal driver of codon usage, whereas points considerably lower than the curve indicate selection pressure as the dominant evolutionary force. The ENC value for each GC3 is calculated as follows:
The s value represents the GC3s content of each codon.
Second parity rule
According to Chargaff’s second parity rule (PR2), the mononucleotides A = T and G = C in the coding sequences suggest that there is no bias for both natural selection and mutational pressure (Sueoka, 1999a). The PR2 is plotted with AT bias at the third codon position [A3/(A3+T3)] as ordinate against GC bias at the third codon position [G3/(G3+C3)] as abscissa to examine the influence of natural selection and mutational pressure on the codon usage pattern. The origin point (0.5, 0.5), at which the coordinates A = T and G = C are located, indicates no bias for both natural selection and mutational pressure. When the value is more than 0.5, it is noticed that purine is preferred over pyrimidine (Sueoka, 1999b). The bias introduced by mutational pressure and natural selection assists in determining the degree of deviation from PR2. In other simple terms, a value greater or less than 0.5 displays a bias caused either by natural selection or mutational pressure or both.
Dinucleotide frequency
The Emboss compseq tool available at https://www.bioinformatics.nl/cgi-bin/emboss/compseq computed the dinucleotide bias, which can affect codon usage in viruses and other organisms. The bias is measured as the odds ratio (Pxy) of observed over expected frequency (Karlin et al., 1994). Typically, values above 1.23 suggest overrepresentation and values below 0.78 suggest underrepresentation. The formula for calculation is:where; fxy, f x, f y represent the frequency of dinucleotide xy, and the frequency of nucleotide x, y respectively.
Phylogenetic analysis
After curating and editing all retrieved whole genome sequences (n = 174) using Bioedit software, each preserving a size of 5500kb, they were subjected to Multiple Sequence Alignment (MSA) via MAFFT software available at https://www.ebi.ac.uk/Tools/msa/mafft. Redundant strains were removed using Jalview software version 2.11.2.5 available at https://www.jalview.org/download/windows/, followed by the detection of putative recombination sites among strains with the Recombination Detection Program (RDP4, version 4.101) available at http://web.cbio.uct.ac.za/∼darren/rdp.html. Putative recombination sites in aligned sequences detected by at least three different default methods (RDP, GENECONV, CHIMAERA, MAXCHI, BOOTSCAN, SISCAN, 3SEQ) were selected. A p-value less than 0.05 determined the statistical significance. The retained sequences (n = 54) underwent phylogenetic analysis after these meticulous processes. The phylogenetic tree was estimated with MEGA software version 11 using a Tamura-Nei model and the Maximum Likelihood (ML) method. An initial tree was inferred by maximum parsimony and tested for phylogeny with 100 bootstraps. Other parameters remained at default settings. The resulting tree, saved in NWK format, was imported to iTOL (https://itol.embl.de/tree/) for annotation and restructuring.
Software and statistical analysis
Spearman’s rank correlation was computed using R software to identify significant factors influencing codon usage patterns. A p-value of <0.01 was considered highly significant, while a p-value of 0.01 < p < 0.05 indicated a significant correlation. The Kruskal-Wallis test evaluated the significance between groups in box plots at 0.05. Additionally, a correlogram was generated using Past software version 4.03, available at https://past.en.lo4d.com/window. Based on the calculated RSCU values of 59 codons for each ASFV strain/lineage, cluster analysis (depicted as a heat map) was performed using the online tool CIMminer6, available at https://discover.nci.nih.gov/cimminer/.
Results
Nucleobase composition
The overall nucleotide frequencies of ORFs, including A%, T%, G%, and C%, were calculated, with results presented in Supplementary Table S2. Other frequency calculations included nucleotide at the third codon position (A3, T3, G3, and C3), overall GC content frequency, average frequency of G + C at the first and second codon positions (GC12), and average frequency of G + C at the third codon position (GC3). A% exhibited the highest overall nucleotide frequency (33.77 ± 0.86), followed by T% (32.08 ± 0.80), with the lowest being C% (17.45 ± 1.01). However, G% (18.44 ± 0.52) displayed minimal variation among the strains. The nucleotide frequency at the third codon position mirrored the overall nucleotide content in a slightly different pattern. The overall %GC content was 35.95 ± 1.35, whereas the GC12 and GC3 contents were 35.96 ± 1.05 and 35.95 ± 2.10, respectively. Furthermore, GC showed a significant correlation with GC12 and GC3, and these variables (GC12 & GC3) also correlated with each other (Figure 12; Supplementary Table S3). Nucleobase composition, GC content, and its counterparts demonstrated a significant correlation with ENC, highlighting their influence on codon usage bias. The prominent role of GC content in codon usage indicates that mutational pressure is the key factor.
Codon usage patterns
Relative Synonymous Codon Usage (RSCU) analysis highlighted variation in codon usage among the strains (Table 1), leading to the emergence of geographically distinct lineages later confirmed by Principal Component Analysis (PCA). Across all lineages, 26 codons exhibited positive bias (with 5 overrepresented), and 26 codons showed negative bias (with 9 underrepresented). The remaining 7 codons displayed slightly uneven expression across lineages (indicated in green), with 4 being negatively biased and 3 showing no apparent bias.
TABLE 1
| Lineage | Host | ||||||
|---|---|---|---|---|---|---|---|
| Aminoacids | Codons | East African | South African | European | Asian | Avarage | Sus scrofa domesticus |
| Phe | TTT | 0.97 | |||||
| Phe | TTC | ||||||
| Leu | TTA | ||||||
| Leu | TTG | 0.85 | 0.88 | 0.89 | 0.81 | 0.86 | 0.64 |
| Leu | CTT | 0.62 | |||||
| Leu | CTC | ||||||
| Leu | CTA | 0.9 | 0.82 | 0.85 | 0.87 | 0.86 | |
| Leu | CTG | 0.74 | 0.75 | 0.76 | 0.7 | 0.74 | |
| Ile | ATT | ||||||
| Ile | ATC | 0.7 | 0.76 | 0.71 | 0.73 | 0.73 | 0.63 |
| Ile | ATA | 0.96 | 0.98 | ||||
| Val | GTT | ||||||
| Val | GTC | ||||||
| Val | GTA | ||||||
| Val | GTG | 0.96 | |||||
| Ser | TCT | ||||||
| Ser | TCC | 0.93 | |||||
| Ser | TCA | 0.76 | 0.79 | 0.81 | 0.82 | 0.80 | |
| Ser | TCG | ||||||
| Ser | AGT | 0.91 | 0.9 | 0.94 | 0.95 | 0.93 | |
| Ser | AGC | 0.92 | 0.87 | 0.87 | 0.95 | 0.90 | 0.81 |
| Pro | CCT | ||||||
| Pro | CCC | ||||||
| Pro | CCA | 0.94 | |||||
| Pro | CCG | ||||||
| Thr | ACT | 0.9 | 0.95 | 0.9 | 0.82 | 0.89 | |
| Thr | ACC | 0.87 | |||||
| Thr | ACA | ||||||
| Thr | ACG | 0.69 | 0.62 | 0.69 | |||
| Ala | GCT | ||||||
| Ala | GCC | ||||||
| Ala | GCA | ||||||
| Ala | GCG | ||||||
| Tyr | TAT | 0.93 | |||||
| Tyr | TAC | 0.68 | 0.64 | 0.62 | |||
| His | CAT | ||||||
| His | CAC | ||||||
| Gln | CAA | ||||||
| Gln | CAG | 0.65 | 0.63 | 0.64 | 0.68 | 0.65 | |
| Asn | AAT | ||||||
| Asn | AAC | 0.81 | 0.82 | 0.79 | 0.79 | 0.80 | |
| Lys | AAA | ||||||
| Lys | AAG | ||||||
| Asp | GAT | ||||||
| Asp | GAC | 0.67 | 0.62 | 0.71 | 0.69 | 0.67 | 0.76 |
| Glu | GAA | ||||||
| Glu | GAG | 0.65 | 0.62 | 0.62 | 0.61 | 0.63 | |
| Cys | TGT | 0.74 | |||||
| Cys | TGC | 0.63 | 0.61 | 0.62 | |||
| Arg | CGT | ||||||
| Arg | CGC | ||||||
| Arg | CGA | 0.97 | 0.91 | 0.85 | |||
| Arg | CGG | 0.83 | 0.76 | 0.85 | 0.77 | 0.80 | |
| Arg | AGA | ||||||
| Arg | AGG | 0.87 | 0.88 | 0.91 | 0.93 | 0.90 | |
| Gly | GGT | 0.85 | 0.88 | 0.92 | 0.96 | 0.90 | 0.71 |
| Gly | GGC | 0.72 | 0.75 | 0.75 | 0.64 | 0.72 | 0.76 |
| Gly | GGA | ||||||
| Gly | GGG | 0.77 | 0.63 | 0.65 | 0.69 | 0.69 | 0.62 |
Mean RSCU values of each codon for the 59 codons in 54 strains (10 ORFs each) were averaged into lineages corresponding to their geographical distribution. In addition, the column values for the host were included for comparison. A codon is said to have a positive codon usage bias if its RSCU value is >1.0, and a negative codon usage bias if its RSCU value is <1.0. Codons with RSCU values <0.6 are considered under-represented, whereas those with values >1.6 are over-represented. Codons with RSCU values of 1.0 are considered unbiased, and used at an equal frequency. Different colors were used to signify codon usage variations: red=positive bias, overrepresented; blue=positive bias, represented; straw=negative bias, underrepresented; black=negative bias, represented; green=average values of the codons expressed uneven across lineages. When compared to the host, the lineages' codon usage appeared to be predominantly antagonistic, with just sporadic instances of concordance. Principal component analysis eliminated codons that encodes for a single amino acid such as Methionine (Met) and Tryptophan (Trp), as well as stop codons.
Interestingly, 22 of the 26 positively biased codons ended in A or T (9 A-ended, 13 T-ended), indicating ASFV’s preference for A/T-ended codons over G/C-ended ones. The five overrepresented codons (RSCU>1.6) were all A/T-ended (TTT, TTA, TCT, AGA, and GGA), while the nine underrepresented codons (RSCU<0.6) were G/C-ended (TTC, CTC, GTC, TCG, CCG, GCG, CAC, AAG, CGC).
When compared to the host, codon usage appeared largely antagonistic, with only sporadic instances of concordance (Table 1). A heat map (Figure 1) was generated to reveal hidden patterns of codon usage across lineages. Two distinct blocks were observed: a green dominant block (primarily representing GC-ended codons, except TCA, CGA and ACT) and a red dominant block (mainly representing AT-ended codons, except GCC and CCC).
FIGURE 1
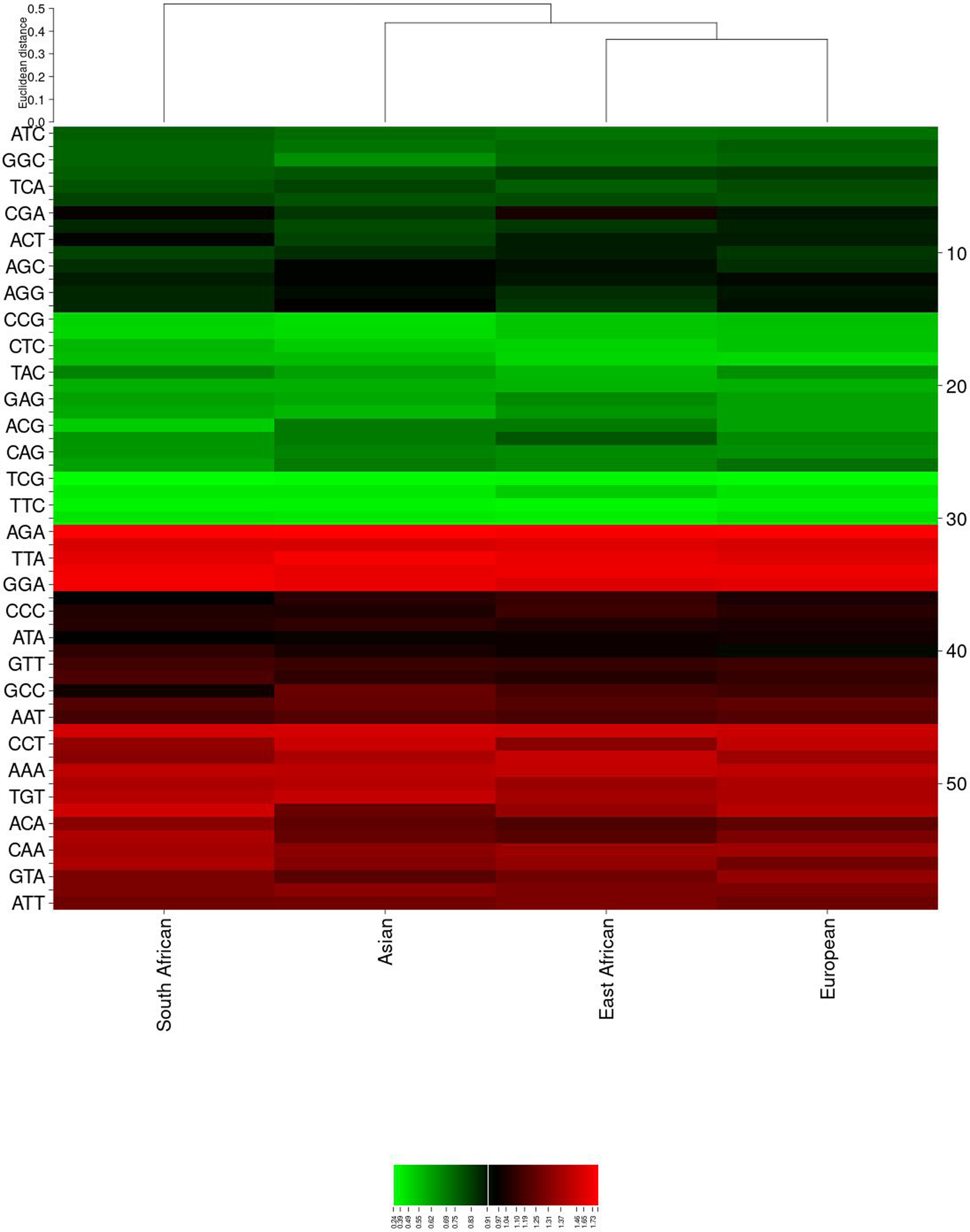
A Heat Map Illustrating RSCU Patterns Based on Lineages. The map delineates four distinct lineages, each represented by a separate column. Rows primarily form two blocks: red for preferred A/T-ended codons and green for non-preferred G/C-ended codons, interspersed with sporadic black patches indicating randomly chosen codons. The RSCU values are arranged into continuous ranges, resulting in observable color variations: green for <1, black for 1–1.6, and red for >1.6. Cluster creation for the CIMminer heatmap analysis employed Euclidean distance and complete-linkage methods.
Upon close examination, it was evident that the green dominant block was a negatively biased cluster of codons (indicating less preferred codons) with blended patterns of sporadic random codon usage. Conversely, the red dominant block represented a positively biased cluster (indicating preferred codons), also interspersed with patterns of random codon usage.
Principal Component Analysis
Principal Component Analysis (PCA) was conducted in R to further reveal the patterns of relative synonymous codon usage. The 59 codon dimensions, each having an RSCU value from specific coding sequences, were condensed into principal components. The first two components (PC1 = 36% and PC2 = 16.9%) accounted for 52.9% of the total variation. The following components, PC3, PC4, and PC5 retained 8.9%, 8.3%, and 5.6% of the variation respectively.
A scree plot was generated to describe how much variation each principal component retained from the original variables. The first four PCs were sufficient to define the necessary variation, but most of the analyses primarily focused on PC1 and PC2 (Figure 2).
FIGURE 2
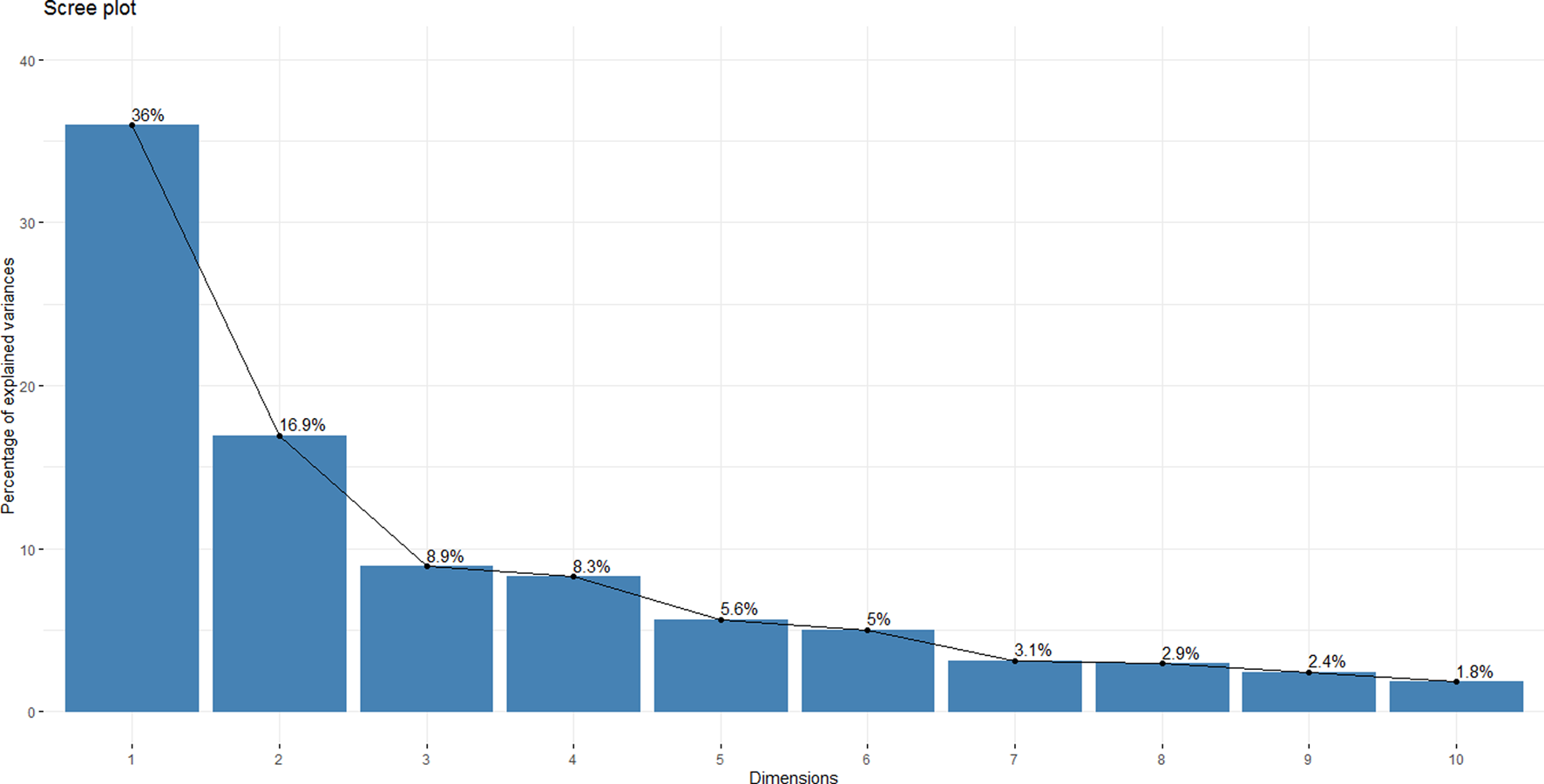
A Scree Plot Indicating Variation Retained by Each Principal Component. The y-axis represents the proportion of explained variation, and the x-axis represents the dimensions of the principal components. The first four components account for 75.7% of the necessary variation, but the analysis primarily engaged the first two principal components, covering 52.9%.
A variable factor map, in the form of a bar graph, was prepared to summarize the top ten most influential variables for each component. Paired component models were also constructed to identify the contribution of two components. Codons TGT, TGC, GAG, and CGC emerged as the most influential in modeling PC1, while TCC, CTC, ACG, and GCC greatly shaped PC2. Similar patterns were observed in PC3, PC4, and in the 1–2, 2–3, and 3–4 component models (Supplementary Figure S1).
A correlating variable plot was computed, revealing significant patterns (Figure 3). On this plot, variables with similar patterns (positive correlation) clustered together, such as those in the same quadrant. Conversely, variables grouped in opposite quadrants demonstrated dissimilar patterns (negative correlation). The right half of the circle revealed G/C-ended codons, while the left half revealed A/T-ended codons. Additionally, the data indicated that G/C-ended codons are negatively correlated to A/T-ended codons.
FIGURE 3
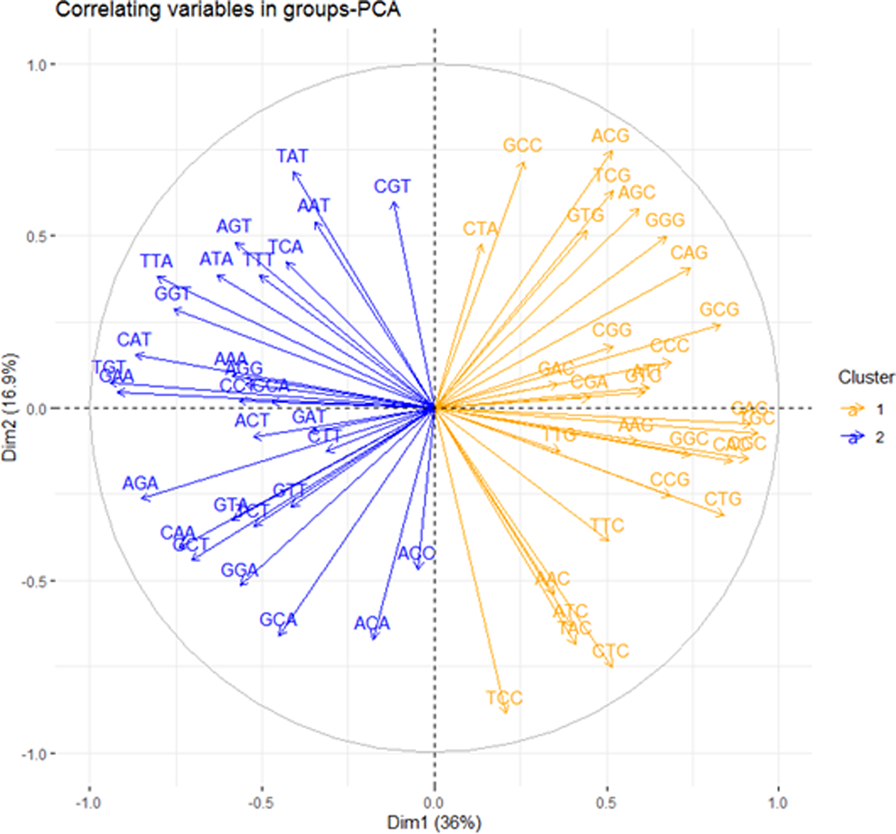
A Variable Correlation Plot. It reveals two distinct clusters: an AT-ended cluster on the left and a GC-ended cluster on the right. Within each cluster, members demonstrate a positive correlation, while members across clusters show a negative correlation. The strength of the correlation between two members is gauged by their angle of inclusion.
Four distinct groups were proposed based on geographical region, namely: Europe, Asia, East Africa, and South Africa. With the available data, we aimed to investigate whether these groups exhibited unique codon usage patterns. Clear, albeit overlapping, clusters emerged, indicating strains/isolates with similar patterns grouping together (Figure 4). Data points were assigned identification numbers for ease of reading. These numbers, alongside detailed strain/isolate specifications, can be found in Supplementary Table S1. For the Asian lineage, strains 48, 15, and 52 showed the highest codon usage patterns. Notably, number 32, displayed to the right of PC1, exhibited extreme codon usage characteristics. For the European lineage, the highest codon usage patterns were observed in data points 40, 45, and 9. In contrast, the East African lineage displayed the highest codon usage in data points 2, 17, 18, 12, 30, 11, 41, 6, and 3, while the South African lineage showed data points 22, 23, 24, 26, and 27 as the highest. All data points that clustered near the origin in each lineage represented average expression, including 37 and 14 for the European lineage, 21 for the Asian lineage, 42 and 43 for the East African lineage, and 5, 7, and 8 for the South African lineage.
FIGURE 4
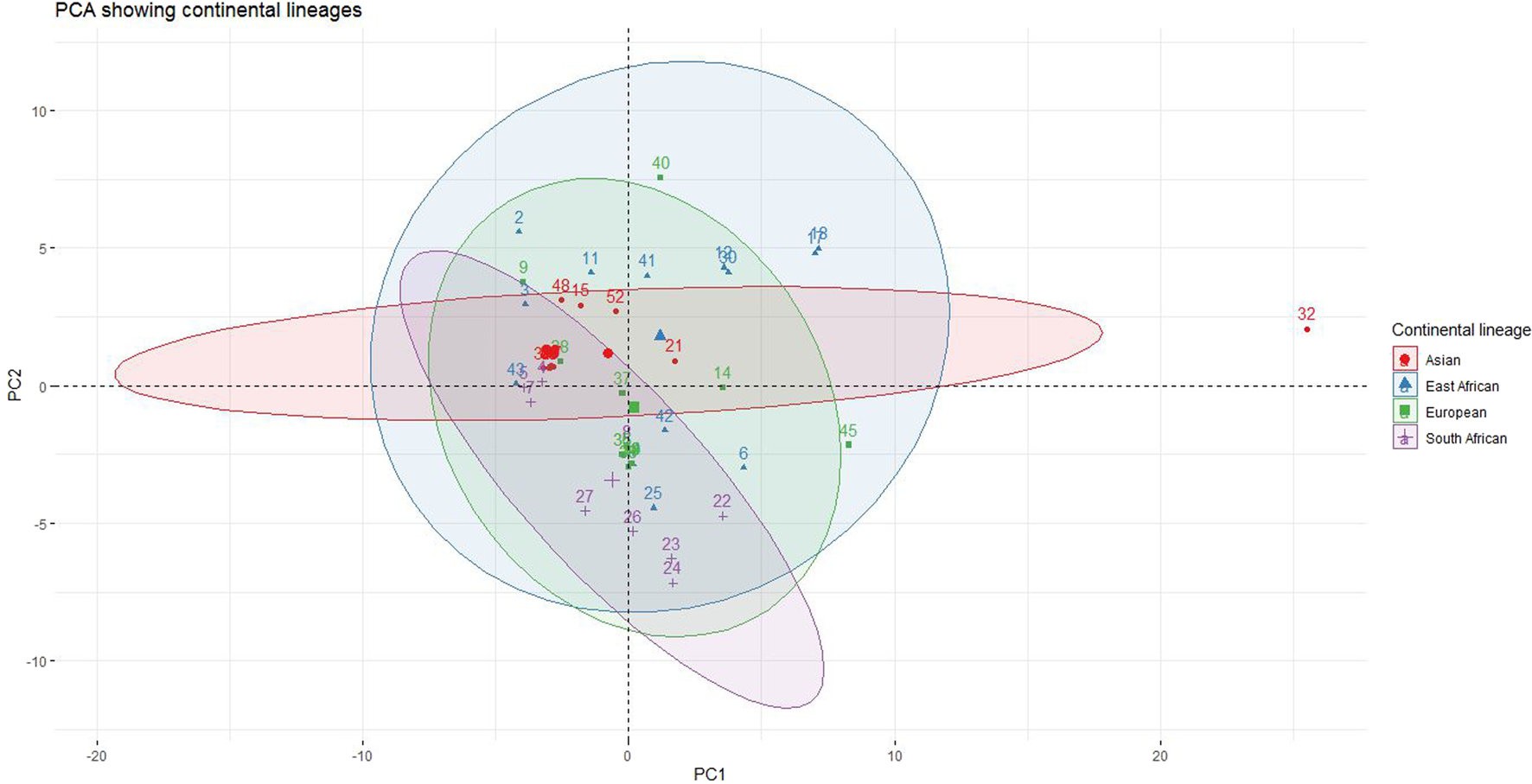
Continental-based Lineages and Their Corresponding Observations. The diagram showcases four lineages, each with observations sharing the background color of the lineage.
Further, a biplot with loading vectors (codons) and individual distributions was prepared to visualize the interrelationships between different loadings and the individual observations they influence, as well as each loading’s correlation with a specific principal component (PC). Clustering was utilized to discern these variations in groups. In essence, an individual on the same side as a given variable was deemed to have a high value for that variable, while an individual on the opposite side of a variable was assumed to have a low value for that variable. Positive loadings indicate a positive correlation between a variable and a PC, whereas negative loadings suggest a negative correlation. Variables covering longer distances (more reddish) from the origin of a PC exert a significant influence on their respective PCs and are well represented on the factor map. Conversely, variables closer to the origin (more bluish) have a lesser influence and are not well represented on that factor map (Figure 5A).
FIGURE 5
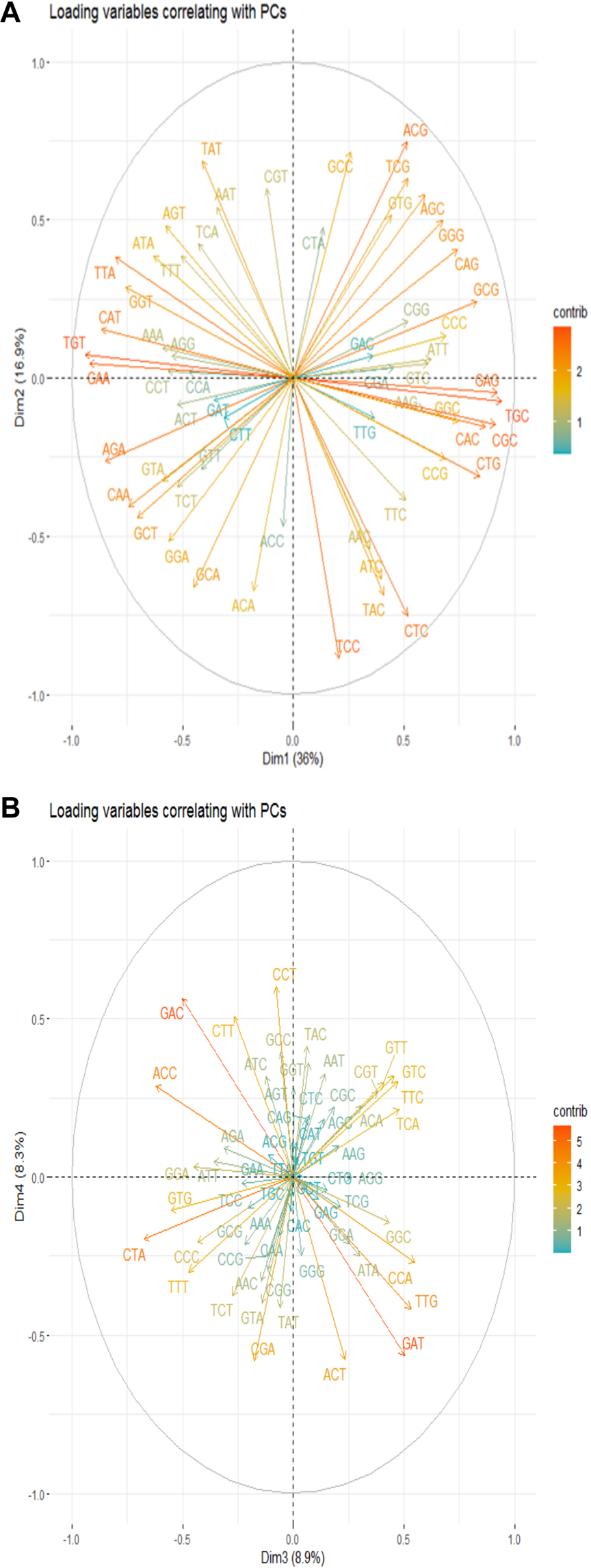
Codon Representation on the PC1 and PC2, and PC3 and PC4 Factor Maps. Arrows of varying lengths and color intensities denote the continuous contribution of loading variables to the principal components.
For instance, variables such as CAG, TGC, CGC, CTG, CAC, and GCG, due to their good representation on the factor map, strongly influence PC1 (positive x-axis), while TGT, GAA, CAT, TTA, GGT, and AGA significantly influence PC1 on the opposite side. Similarly, ACG, TCG, GCC, GTG, AGC, TAT, and AGT, due to their good representation on the factor map, exert more effect on PC2 (positive y-axis) while TCC, CTC, TAC, ACA, and GCA affect PC2 on the opposite side.
Less intense and shorter variables can also be well represented on other PCs. For example, GAT, GAC, TTG, CTA, and ACC variables are underrepresented on the Dim 1-2 correlation circle but well-represented on the Dim 3-4 correlation circle (Figure 5B).
Additionally, the smaller the angle between the variable and a PC, the larger the correlation; conversely, a wider angle between them signifies a smaller correlation and increased non-specificity. With this in mind, all codons forming a positive angle with PC1 (positive x-axis) up to 45° strongly correlate positively with PC1; a hypothesis that similarly applies to PC2 (positive y-axis). The remaining codons in the same quadrants, including an angle from 45° to 90°, have a weak positive correlation with their respective components, and these codons can also be redistributed to other principal components. This hypothesis also applies to the codons taking a negative angle from the origin on the opposite. In summary, codons orienting near the horizontal to PC1 or vertical to PC2 and close to the circumference are the most influential to their respective PCs (as per the examples above).
From this analysis, it appears that a significant part of the individual observations in the East African lineage expresses GC-ended codons (see clusters II and III on the positive PC1-axis and cluster II on the positive PC2-axis, Figures 6A, B). Meanwhile, a significant part of the Asian lineage observations appears to express AT-ended codons (see cluster I on the positive PC2-axis and negative PC1-axis). South African lineage observations feature an expression of AT-ended codons on both negative axes of PCs and GC-ended codons (mostly C-ended codons) on the negative axis of PC2 (see cluster IV for both negative directions of PCs and cluster III on the negative direction of PC2). Finally, much like the East African lineage, a significant part of the European lineage is greatly influenced by a diversity of GC-ended codons (see cluster II and III on the positive PC1-axis and cluster III on the negative PC2-axis).
FIGURE 6
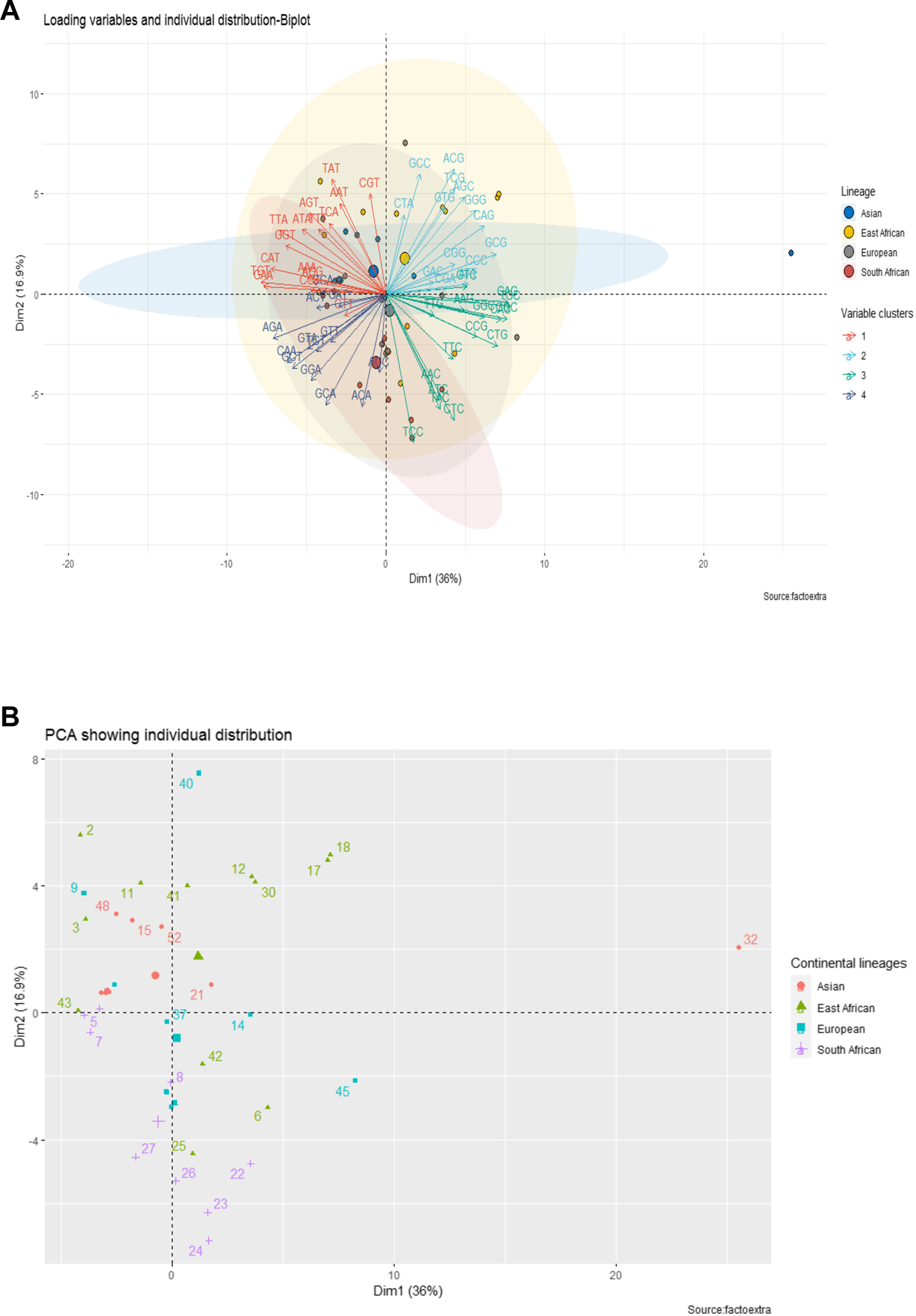
Individual Distribution in Lineages Influenced by Loading Variables. Part A shows individuals and variables clustered together, with the color of individual observations mirroring the background color of the cluster. A variable (codon) in line with an observation has a significant influence on that observation, whereas a variable opposite an observation exerts minimal influence. Part B reveals clustered individuals without background color, acting as a visibility-enhancing counterpart to Part A, particularly highlighting individual distribution in the absence of variables.
Codon usage bias and ENC-GC3 plot analysis
The Effective Number of Codons (ENC) is a prominent metric for calculating Codon Usage Bias (CUB). The average ENC value for all the coding sequences studied is 52.8 (range: 52.8–56.1 ± 0.99), and the CUB, in this case, is generally low (greater than 45). Multiple comparison box plots (Figure 7) revealed significant differences in bias between lineages (p < 0.05).
FIGURE 7
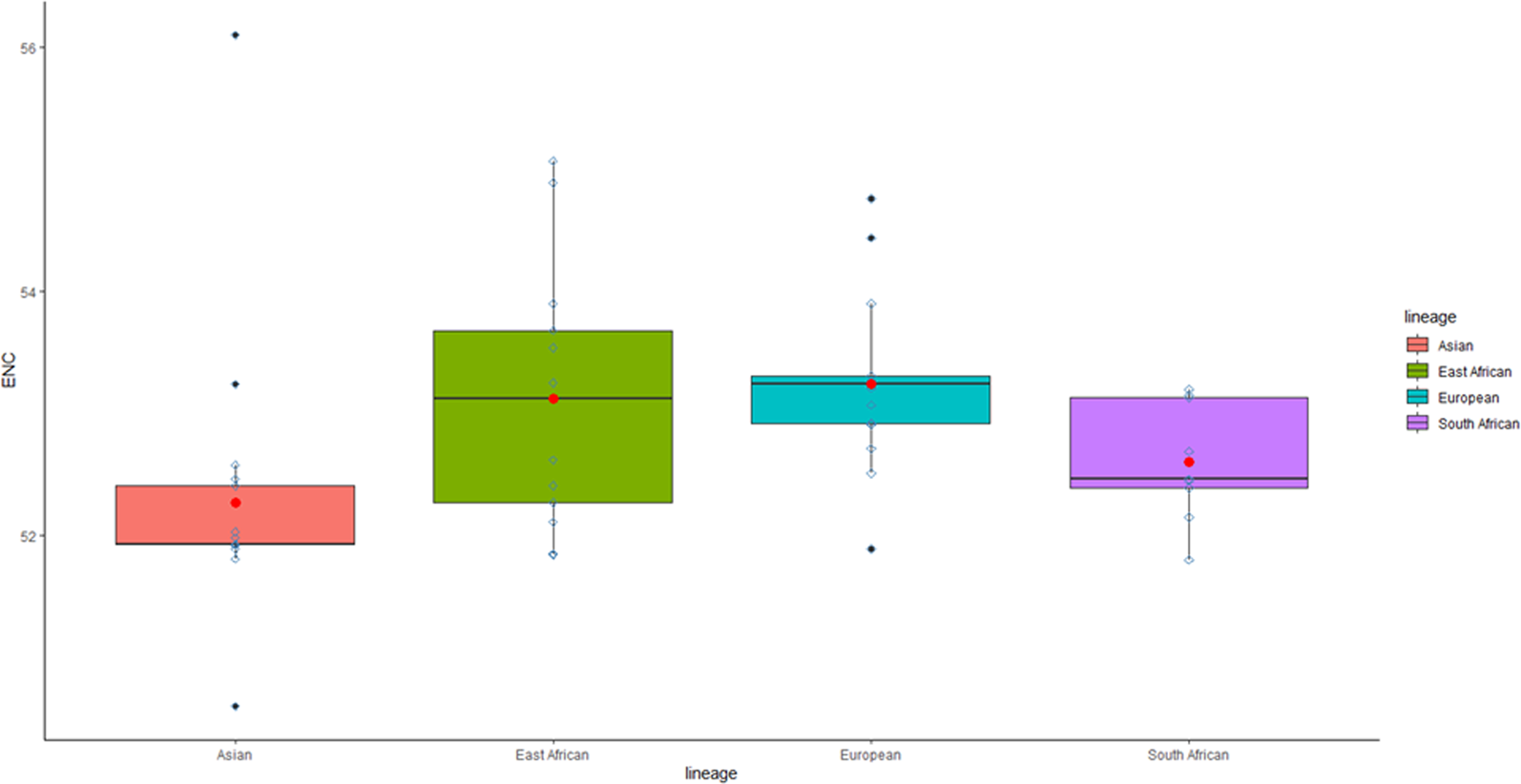
Multiple Comparison Box Plots Depicting ENC. The codon usage bias is evaluated by comparing individual lineages. The box length signifies a 50% proportion of total data, with whiskers indicating the remaining 50%. An upper whisker represents 25% maximum values, and a lower whisker depicts 25% minimum values. The box’s black line denotes the median, the red point represents the mean, and outliers are marked as black dots beyond the whiskers.
The European lineage shows a slightly higher median value (as indicated by the line within the box plot) than the East African lineage, followed by the South African lineage in terms of magnitude. However, the Asian lineage has the lowest median value of all. Consequently, the European lineage demonstrates the least CUB, followed by the East African lineage, while the Asian lineage exhibits the highest CUB.
Contrastingly, the East African lineage displays the most diversity in its codon usage, followed by the European lineage, with the Asian lineage being the least diverse. This diversity is apparent when examining the range (R) and interquartile range (IQR), which respectively measure the spread of the entire dataset and the middle 50% of the data distribution. The East African lineage has both the highest R and IQR, indicating its diverse codon usage. The European lineage has a larger R than the South African and Asian lineages but has a smaller IQR than these two lineages.
This apparent contradiction can be clarified by examining the shape of the data distribution in addition to the data spread. The East African lineage data is negatively skewed, meaning that the lower portion of the box is longer than the upper portion and that the median is closer to the upper end. Similarly, the European lineage’s median is even closer to the upper end, indicating a similar pattern. This pattern is not observed for the South African and Asian lineages, which show a positive skew, meaning the median aligns with or is positioned closer to the lower end.
A negatively skewed distribution suggests the presence of more low-score observations and fewer high-score observations. The high-frequency data points are primarily represented in the upper end, while the low-frequency data points are spread in the lower end. Positively skewed data distribution shows the opposite pattern.
These statistical insights suggest that the East African and European lineages are dominated by consistently high ENC values, which biologically implies low CUB, allowing for more diverse codon usage compared to the South African and Asian lineages. Furthermore, independent of the distribution shape, the box of the European lineage is above the South African and Asian lineages, which can be interpreted as a high ENC score, and consequently, low CUB. This allows a wider range of codon usage.
In conclusion, even though the interquartile range (IQR) of the European lineage, as indicated by the length of the box, was less than that of the South African and Asian lineages, the European lineage still displayed greater codon usage diversity than the South African and Asian lineages. The range (R) and the shape of the data distribution, as statistical measures of group differences, provided additional support for the European lineage, in contrast to the South African and Asian lineages, which were only supported by IQR.
To better understand the primary evolutionary forces at play—natural selection versus mutational pressure - in shaping biased codon usage, we used the ENC-GC3 plot. Mutational pressure is considered the sole factor when individual observations lie on or just below the curve. However, when observations fall significantly below the curve, it indicates that other factors, such as natural selection, may also be contributing. In this study, most observations were found to be well below the curve (Figure 8), suggesting that natural selection may play a crucial role in influencing codon usage bias.
FIGURE 8
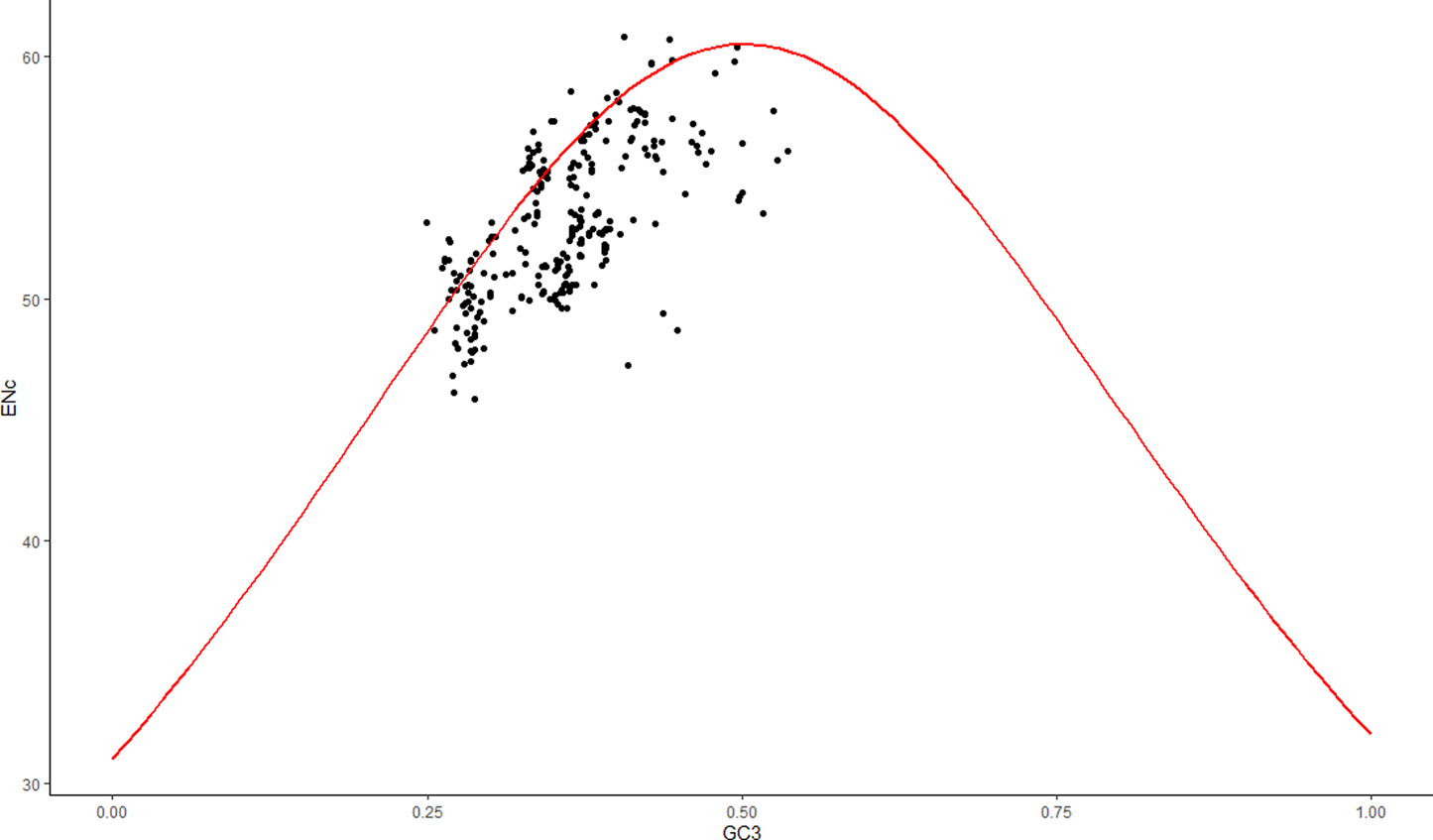
The ENC-GC3 Plot. The red line depicts the expected trend with data points distributed above, on, and below it. The significant underplacement of points away from the curve suggests influences beyond mutational pressure, primarily natural selection, that shape the codon usage bias of ASFV.
Neutrality plot analysis
Further elucidation of the principal influences on codon usage was achieved through neutrality plotting (Figure 9). The regression slope of the GC12 versus GC3 plot nearing 0 suggests the preponderance of natural selection, while a slope approximating 1 signifies the dominance of mutational pressure, indicative of complete neutrality. A robust correlation between GC12 and GC3 (R = 0.74, p < 0.01) initially indicated the supremacy of mutational pressure over natural selection. However, the regression equation’s slope, y = 0.43x+21, delineates the contributions of mutational pressure and natural selection as 43% and 57% respectively, highlighting a slight dominance of natural selection over mutational pressure.
FIGURE 9
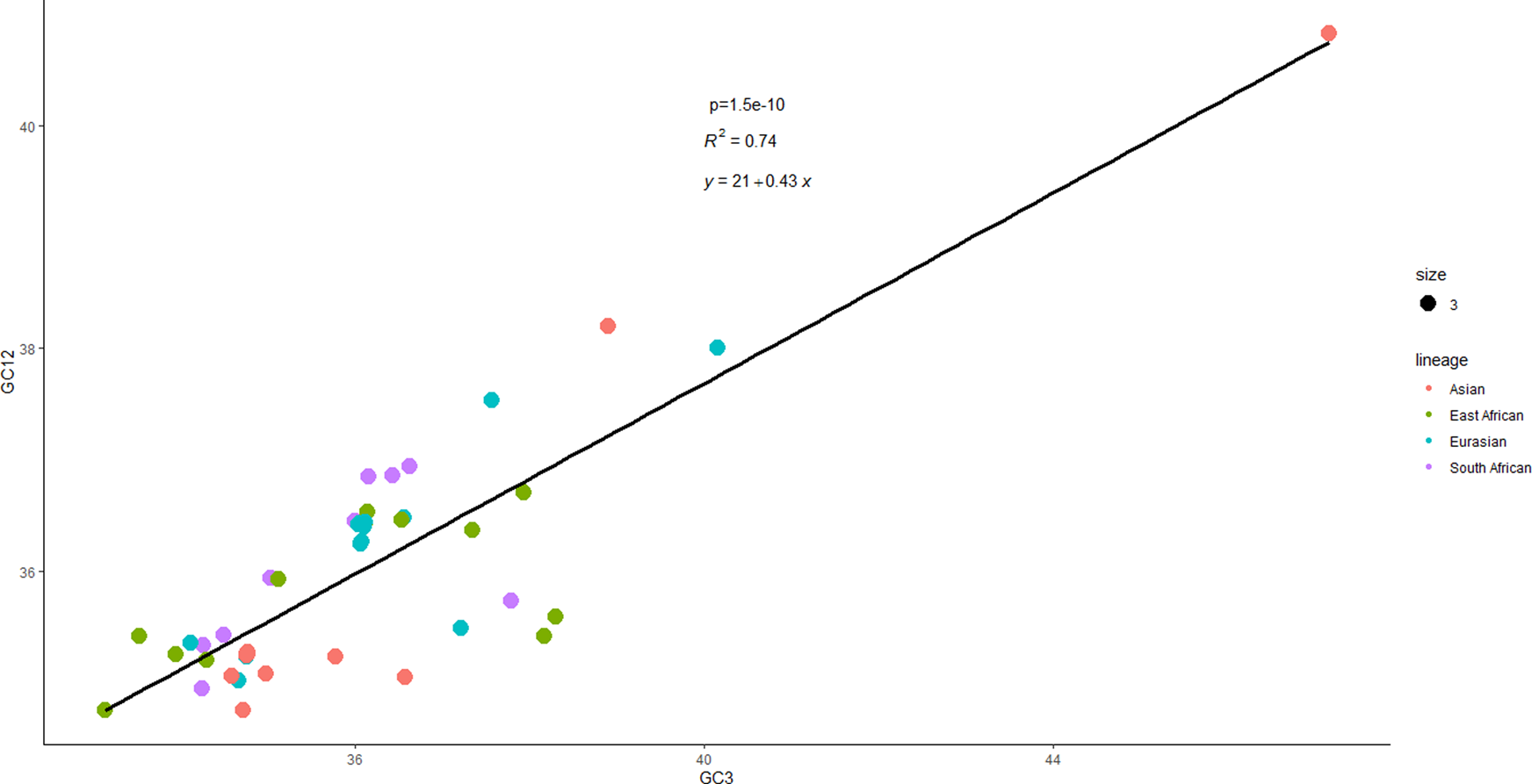
A Neutrality Plot. This regression chart between GC12 and GC3 values discloses a 0.43 and 0.57 proportional contribution to evolution from mutational and natural selection, respectively.
The second parity rule
The construction of the parity graph assumed no bias towards either mutational pressure or natural selection if the origin coordinates were positioned at 0.5, 0.05 (Figure 10). In contrast, the presence of bias, driven by either AT bias, GC bias, or both, would instigate a departure from the PR2 rule where A≠T and G≠C, and purines would be preferred over pyrimidines for values >0.5. The mean values (0.62, 0.35) demonstrated a 1.2-fold increase in AT bias and a 1.5-fold reduction in GC bias from the origin. These findings underscore the predominance of natural selection over mutational pressure, with both acting concurrently.
FIGURE 10
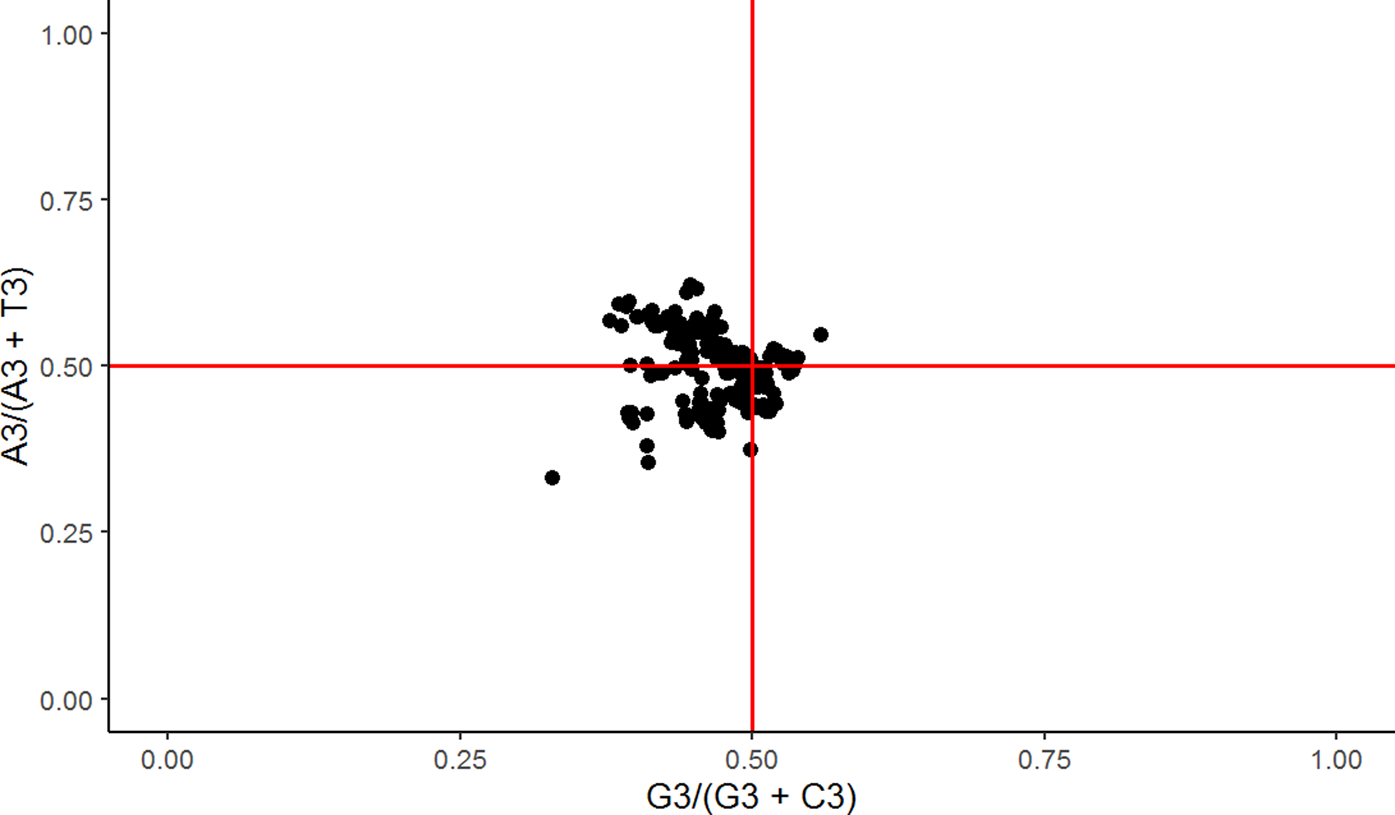
A Parity Plot. The y-axis represents an AT bias, and the x-axis denotes a GC bias. The proportional deviation of data points from the origin (0.5, 0.5) demonstrates a violation of the second parity rule. The AT bias deviates from the origin by an increase of 1.2, while the GC bias deviates by a decrease of 1.5.
Protein biochemical features as determinants of natural selection
The biochemical metrics (Gravy and Aroma scores) assessed the impact of protein biochemistry on natural selection. Distinct lineage differences were clearly portrayed through multiple comparison box plots (Figures 11A, B), highlighting the role of hydropathicity and aromaticity in codon usage bias. The divergence in these parameters across lineages underscores the importance of natural selection in shaping ASFV’s codon usage. Median gravy scores indicate that African lineages, characterized by elongated boxes and elevated medians, exceed Eurasian lineages in terms of increased hydropathicity positivity. Similarly, African lineages, with higher medians, showed a greater inclination towards positive aromaticity compared to Eurasian lineages. This trend suggests the geographical modulation of natural selection, manifesting as continental differences at the protein biochemistry level. Subsequent correlation analysis of the gravy and aroma scores with the first two principal components confirmed a significant correlation, further substantiating the influence of natural selection on codon usage.
FIGURE 11
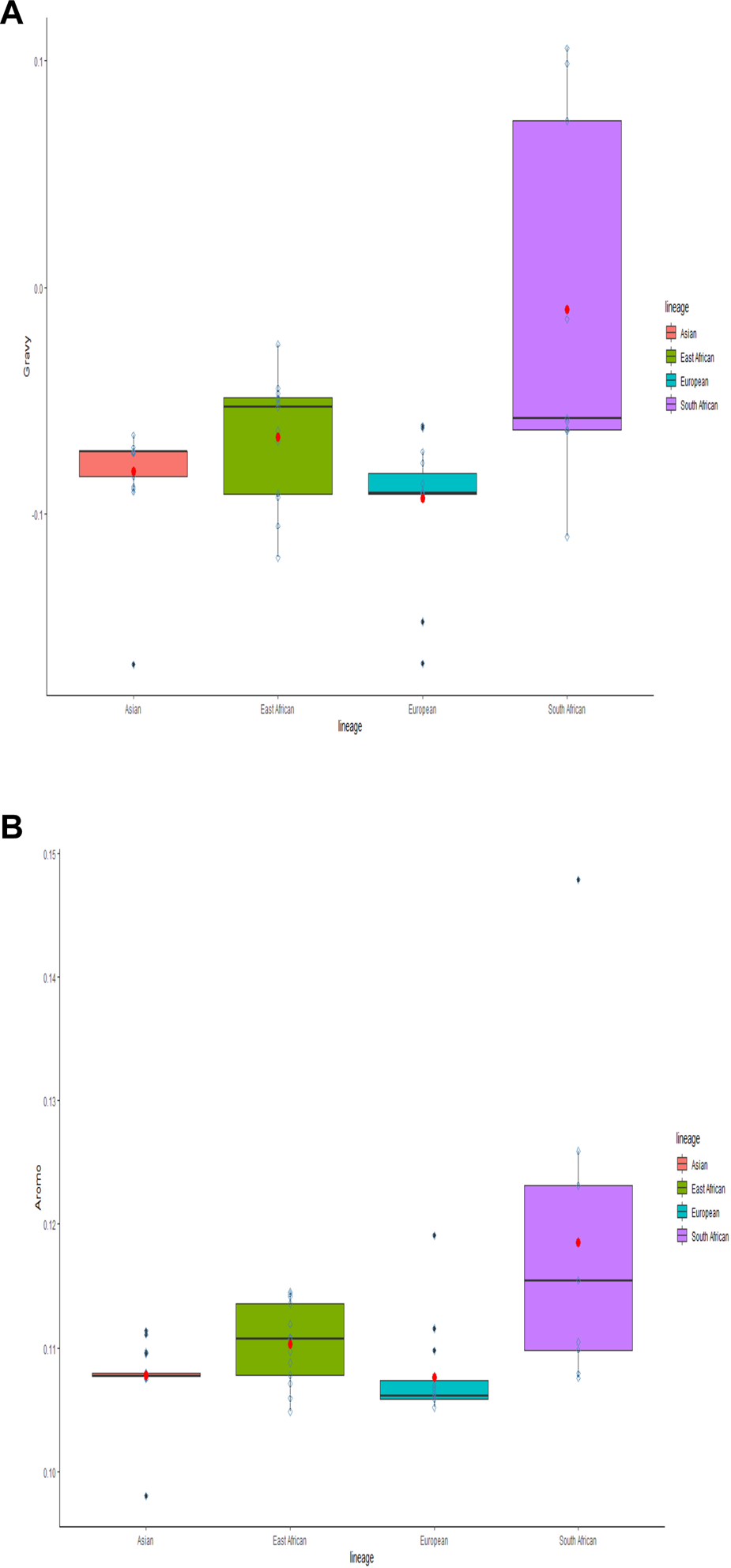
Multiple Comparison Box Plots Displaying the GRAVY and Aroma Scores. The scores are assessed by juxtaposing individual lineages. The box length denotes a 50% proportion of total data, with the whiskers illustrating the remaining 50%. An upper whisker represents 25% maximum values, and a lower whisker shows 25% minimum values. The box’s black line indicates the median, the red point shows the mean, and outliers are represented as black dots beyond the whiskers.
Codon Adaptation Index (CAI)
The Codon Adaptation Index (CAI) was employed to evaluate the adaptation of ASFV genomes to their respective hosts, gauging the influence of natural selection on viral genomic expression. A higher CAI value (0–1) corresponds to greater adaptation. The expected value (eCAI) calculated from the coding sequences yielded 0.598 (p < 0.05), indicating that 60% of the viral codon usage is adaptive. Although ASFV demonstrates real codon adaptation, it manifests as moderate codon expression, with 40% of viral codons unutilized in active replication cycles, potentially expressed during latency phases (Kumar et al., 2018). Correlation analysis with PC1 and PC2, akin to other codon usage parameters, revealed a significant correlation, verifying ASFV adaptation. Furthermore, a correlation analysis between CAI and ENC was conducted to validate this adaptation as a result of natural selection’s influence on CUB.
Relative Codon Deoptimization Index
The RCDI established codon usage similarity at the gene level. Unlike CAI, a higher RCDI value (>1) signifies codon use dissimilarity (codon deoptimization and reduced adaptability) relative to the host. Conversely, an RCDI value close to 1 indicates codon use similarity (codon optimization and viral adaptability). The expected RCDI value derived from this study was 3.003 (p < 0.05), denoting that the virus’s codon expression diverges from the host’s, implying a low to moderate level of adaptation. In other words, ASFV chooses not to express certain codons during gene expression, thus surrendering that codon usage gap to the host. This codon deoptimization strategy avoids host competition and somewhat restricts viral translation speed, thereby controlling viral replication rate (Butt et al., 2014; Butt et al., 2016). This offers a unique advantage by moderating translation speed to ensure optimal protein folding for effective replication cycles. The virus may selectively utilize codons that the host does not prefer for its survival, fitness, and immune evasion (Butt et al., 2014; Butt et al., 2016; Yao et al., 2020).
Similarity index analysis
Beyond the aforementioned adaptation indices, we sought to ascertain the influence of the host on overall codon usage in viral gene expression. The Similarity Index (SiD) provided this perspective. A higher value, nearing 1 (range, 0–1), signifies a strong overall impact of the host on viral codon usage patterns. The host impacted ASFV codon usage patterns by approximately 50% (0.49), implying that around half of the viral codons in any gene expression remain unaffected by the host.
Correlation analysis
Autocorrelation was conducted to ascertain the association among nucleotide base composition parameters, ENC, CAI, GRAVY, and Aroma scores, and the first two principal components. The aim was to identify a significant relationship between these indices and PC1 and PC2. Additionally, the length of the open reading frame (ORF) was considered to validate the hypothesis that gene length influences codon usage patterns. The first two components were chosen as they encapsulated a substantial proportion of CUB (52.9%). The results (Supplementary Table S3; Figure 12) portrayed varied correlation patterns among parameters at different significance levels (0.001<p< 0.05). The correlogram showed a significant correlation between the overall nucleotide base composition and the nucleotide base at the third codon position with either PC1 or PC2 (0.001<p< 0.05), implying that nucleotide compositional constraint is a determinant of CUB. A highly significant positive correlation (p < 0.01) was observed between ENC and PC1, with a similar positive correlation noted between CAI and PC1. Conversely, the Gravy and Aroma scores displayed a significant correlation (p < 0.05) with PC2. The length of the coding sequences was positively correlated with both PC1 and PC2, highlighting the importance of gene length in CUB. Additionally, a variable correlation plot was drafted to examine the correlation between loading vectors, in this case, codons (Figure 3).
FIGURE 12
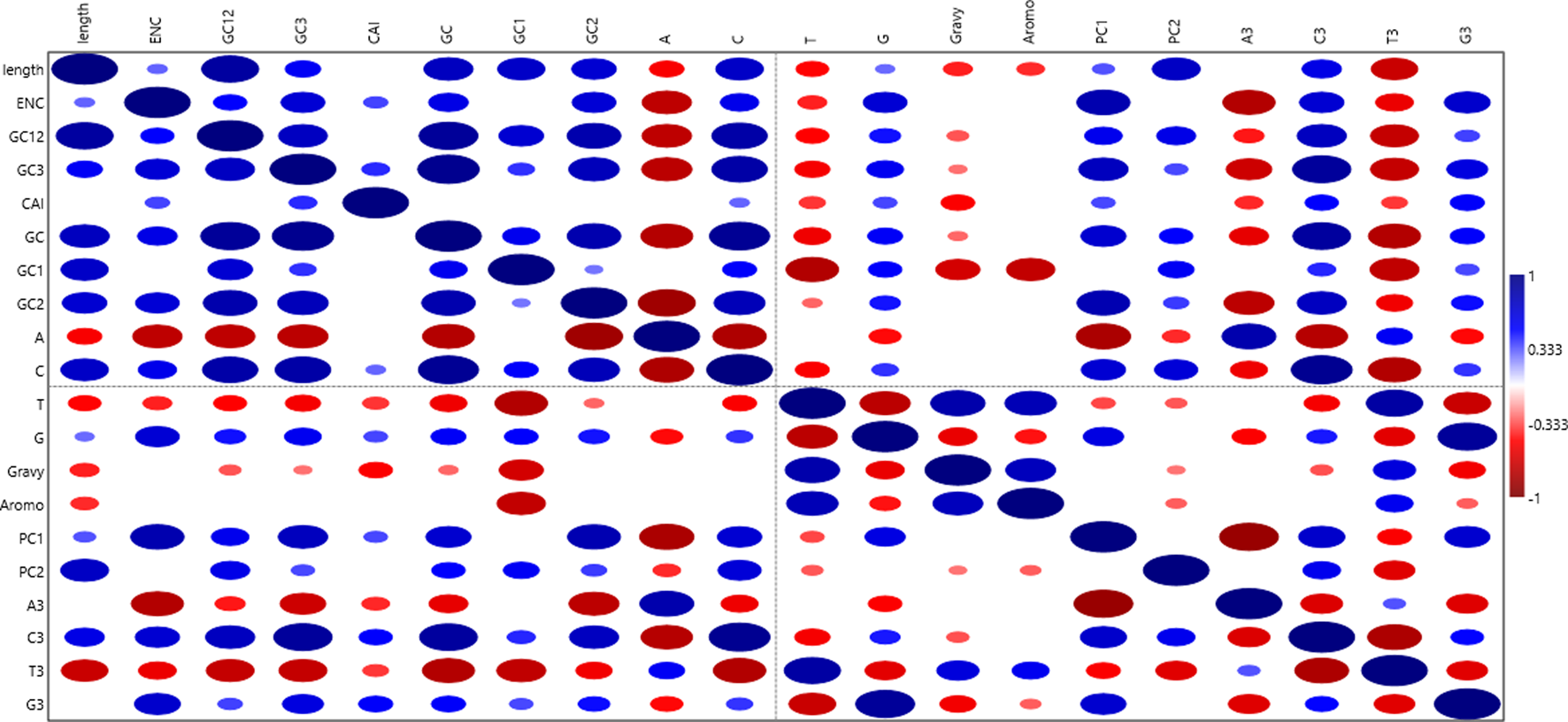
Correlogram Illustrating Autocorrelation Among Variables. The cycle’s relative size represents the correlation strength, mirrored by various color intensities. Both positive and negative correlations are depicted continuously, ranging from a maximum (+1) for a strong positive correlation to a minimum (−1) for a strong negative correlation. Areas without cycles suggest the absence of correlation between the variables.
Dinucleotide frequency analysis
The dinucleotide frequency analysis estimated the relative abundance of 16 dinucleotides in ASFV’s coding sequences. None of the dinucleotides were at their expected frequency or randomly distributed (Table 2). The results indicated an over-representation of dinucleotides AA, TT, AT, and TA (ρxy >1.23), whereas underrepresented dinucleotides included CG, CC, GC, GG, and GT (ρxy<0.78). To confirm this, eight codons featuring the dinucleotide CpG (TCG, CCG, ACG, GCG, CGT, CGC, CGA, and CGC) from the earlier RSCU values were reassessed. Four of these (TCG, CCG, GCG, CGC) were underrepresented, verifying the presence of dinucleotide usage bias. Interestingly, the dinucleotide TρA, usually found to be underrepresented in many studies, was slightly overrepresented in this research (ρxy = 1.356) (Cheng et al., 2013). This was supported by the presence of TTA among the five overrepresented codons (TTT, TTA, TCT, AGA, and GGA) in the RSCU values. Furthermore, an influence from the positive AT bias in nucleotide composition contributed to the overrepresentation of the dinucleotide TρA in ASFV. The dinucleotide TT was overrepresented in the RSCU values of the TTT and TTA codons, whereas the dinucleotides AA and AT were not overrepresented.
TABLE 2
| Dinucleotide | East African | South African | European | Asian | Average | STD |
|---|---|---|---|---|---|---|
| AA | ||||||
| AC | 0.834 | 0.860 | 0.856 | 0.837 | 0.847 | |
| AG | 0.886 | 0.849 | 0.902 | 0.911 | 0.887 | |
| AT | ||||||
| CA | 0.946 | 1.038 | 0.981 | 0.989 | 0.989 | |
| CC | ||||||
| CG | ||||||
| CT | 0.898 | 0.917 | 0.892 | 0.874 | 0.895 | |
| GA | 1.049 | 0.971 | 1.066 | 1.062 | 1.037 | |
| GC | ||||||
| GG | ||||||
| GT | ||||||
| TA | ||||||
| TC | 0.782 | 0.847 | 0.785 | 0.793 | 0.802 | |
| TG | 1.056 | 1.061 | 1.062 | 1.046 | 1.056 | |
| TT |
None of the dinucleotides were distributed randomly or at their predicted frequency. Values above 1.23 (ρxy >1.23) and below 0.78 (ρxy <0.78) indicate overrepresentation and underrepresentation, respectively. The variance in dincucleotide usage is indicated by colors. Black = represented, Blue = underrepresented, and Red = overrepresented. STDs are denoted by maroon.
Phylogenetic analysis
The evolutionary relationships of ASFV’s coding sequences, based on geographical lineage, were reaffirmed through the construction of a phylogenetic tree (Figure 13A). The lineages were grouped into Asian, European, East African, and South African clades. Interestingly, however, some strains from different clades were found nested within foreign clades; this could be partly attributed to genotypic relationships between some of the strains and the parent clades where they originate.
FIGURE 13
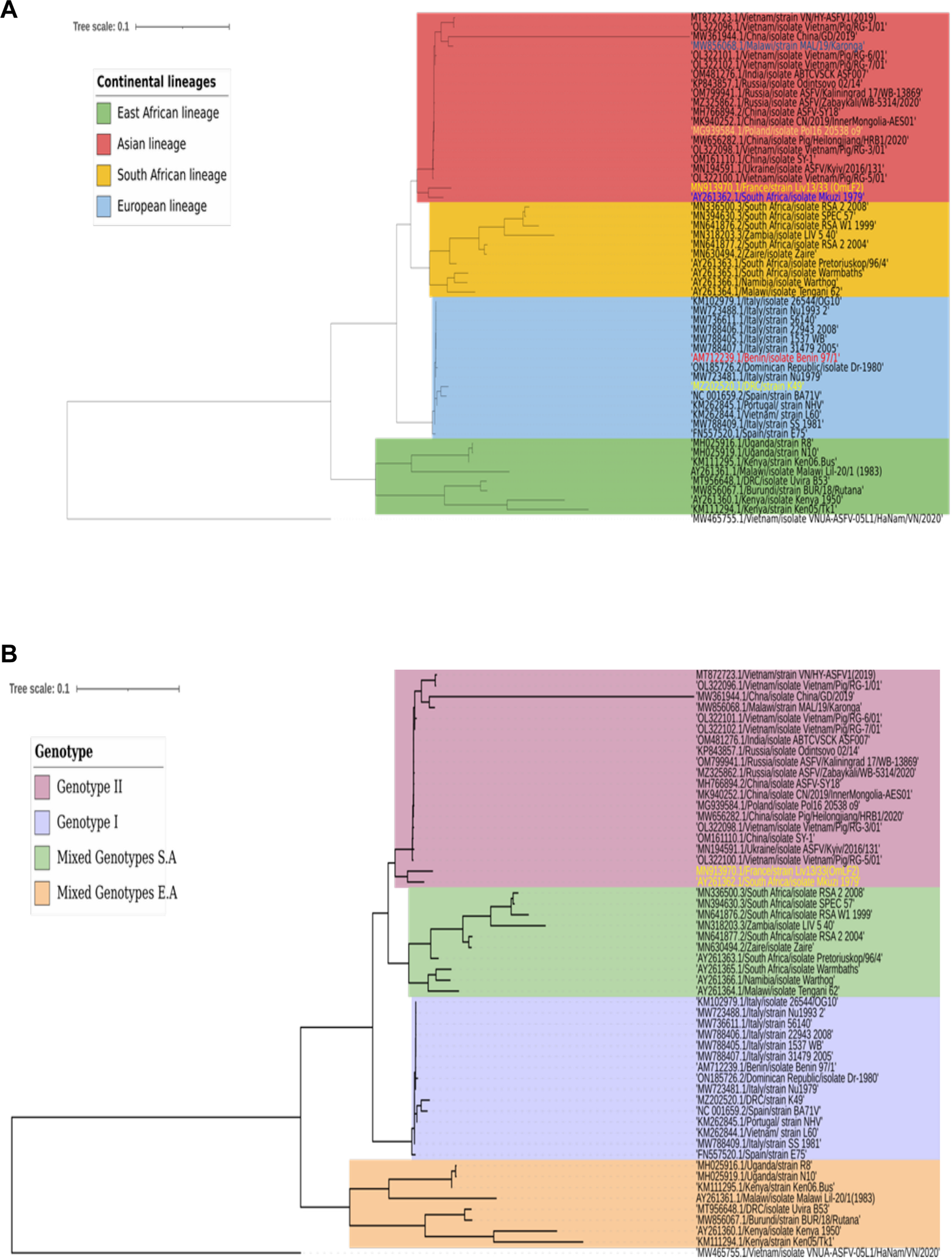
A Maximum Likelihood (ML) Phylogeny. The tree was constructed using the Tamura-Nei model and processed with 100 bootstraps in MEGA 11 software for about 5 hours. The tree, saved in NWK format, was imported to iTOL online software for annotation. Isolates with analogous characteristics are grouped into a single clade, while those with distinct features form different clades. Tree A reveals the evolutionary relationships of the continental lineages, with some isolates nested in foreign clades, identified by color differentiation from the background. Tree B demonstrates the evolutionary relationships of the genotypic lineages, with most nested isolates from Tree A absent, indicating a genotypic relationship independent of geographic distribution.
This observation is supported by a phylogenetic tree constructed based on genotypes, where nesting was eradicated except for the French strain (MN913970.1/strain Liv13/33 (OmLF2) and South African strain (AY261362.1/isolate Mkuzi 1979) (Figure 13B). These strains were found nested in the genotype II clade even though they are genotype I members. All these observations elucidate the complex transmission chains and evolutionary dynamics of ASFV in nature.
Discussion
The nucleotide and codon usage composition showed a significant correlation with ENC, indicating its influence on CUB (Ma et al., 2017; Gu et al., 2020). The GC content (35.95%) characterizes ASFV as an AT-rich DNA virus. ASFV’s preference for using AT nucleotides over GC nucleotides in all its coding sequences has been observed in a comparative genomics analysis of ASFV (Pu et al., 2023). Other viruses with a similar AT nucleotide composition include Hantaan viruses (Ata et al., 2021), Hepadnaviridae (Deb et al., 2020), Flaviviridae (Yao et al., 2019), lyssaviruses (Zhang et al., 2018), PEDV (Chen et al., 2014), PDCoV (He et al., 2019), and SARS-CoV-2 (Hou, 2020).
The significant correlation of GC content with ENC suggests that mutational pressure is the evolutionary force driving the observed biased codon usage (Zhou et al., 2013; Deb et al., 2021b). However, the weak to moderate correlation of nucleotide content and codon composition with the first two principal axes suggests that other factors, such as natural selection, are also in play alongside mutational pressure.
The ENC, an absolute measure of CUB, was 52.80, indicating that ASFV has a low codon usage bias (>45) (Hemert et al., 2016). This result aligns with those for PDCoV (ENC = 52.69) (Peng et al., 2022), SARS-CoV-2 (ENC = 45.38) (Hou, 2020), PEDV (ENC = 47.91) (Chen et al., 2014), MERS-CoV (ENC = 49.82) (Chen et al., 2017), and other viruses.
The low biased codon usage value suggests that ASFV uses a wide range of its synonymous codons within a respective codon family for efficient translation and subsequent productive viral replication cycles (Hemert et al., 2016). The significant difference in lineages’ENC shown by box plots (Figure 7) supports the hypothesis that natural selection under geographical or environmental influence, in addition to mutational pressure, accounts for the observed viral evolutionary differences. Furthermore, ENC-GC3, PR2, and neutrality plots demonstrated that both natural selection and mutational pressure are major drivers for evolution in ASFV. However, the neutrality plot analysis revealed that natural selection is somewhat more influential than mutational pressure by 14%.
Codon usage patterns, as indicated by RSCU, revealed 22 out of 26 positively biased codons (RSCU>1) as A/T-ended. Unsurprisingly, all five overrepresented codons (RSCU>1.6) were also A/T-ended. Conversely, all nine underrepresented codons (RSCU<0.6) were G/C-ended. RSCU values also uncovered lineage-based codon usage patterns, a finding further substantiated by PCA. The striking contrast between the codon usage expression of the viral lineages and that of the host suggests that ASFV experiences lower translation efficiency, thereby allowing proteins to fold properly during translation (Table 1). These findings suggest that natural selection has a greater impact than mutational pressure.
The heat map representation of RSCU lineage patterns (Figure 1) presented two distinct codon blocks: an A/T-ended block and a G/C-ended block. It was observed that the A/T-ended block was highly expressed (RSCU>1), while the G/C-ended block was least expressed (RSCU<1), which reiterates the dominance of A/T-ended codons in ASFV.
PCA identified four clusters, each corresponding to geographically-oriented lineages with individual observations. However, these clusters overlap one another suggesting that mutational pressure is a common evolutionary force at play among strains/isolates, regardless of their geographical origins. High codon usage patterns within clusters signal strains/isolates with pronounced CUB in each region. Interestingly, lineages were seen to favor GC-end, AT-end, or both, in their codon usage. For instance, the East African and European lineages skewed heavily toward G/C-ended codons, while the Asian lineage favored A/T-ended codons. Only the South African lineage seemed to be influenced by both codon ends. East African lineage displayed the most diverse codon usage, closely followed by the European lineage, as indicated by the detailed CUB analysis using ENC and supported by PCA. Conversely, the Asian lineage appeared to show the least codon usage diversity. These findings suggest that, in addition to mutational pressure, natural selection likely plays a significant role in ASFV evolution.
GRAVY and Aroma scores indicated significant differences among geographically-based lineages (p < 0.05). These scores further support the influence of natural selection as a fundamental force in ASFV evolution (Chen et al., 2014; Yu et al., 2021b). This was especially apparent when multiple comparison box plots showed the divergence of African and Eurasian lineages in terms of their hydropathicity and aromaticity tendencies. As indicators of natural selection, these scores also positively correlated with PC2, providing additional evidence that natural selection slightly exceeds mutational pressure.
Indicators of adaptation such as the Codon Adaptation Index (CAI, eCAI = 0.598), Relative Codon Deoptimization Index (RCDI, 3.003>1), and Similarity Index (SiD, 0.49) all support the notion that ASFV is co-adapting with its host in some way. These metrics demonstrate that ASFV has moderately adaptive features and has managed to deoptimize its codons to some extent. Notably, the Similarity Index indicates that ASFV prefers to express 50% of its codons to reduce competition with the host during active replication cycles (Butt et al., 2014; Butt et al., 2016). This antagonistic codon usage, also observed in hepatitis A virus, shrimp viruses, and chikungunya virus (Andrea et al., 2011; Butt et al., 2014; Tyagi et al., 2017), suggests that ASFV is not highly host-specific, given its ability to infect hosts other than domestic pigs, such as warthogs that serve as reservoirs for maintaining sylvatic cycles (Lubisi et al., 2005; Costard et al., 2009; Brown and Bevins, 2018; Tian et al., 2018; Craig et al., 2021). Additionally, the CAI of 60% supports the recurrent assertion of natural selection’s influence (Kumar et al., 2018).
The marked decrease in CpG dinucleotides may be a response to host antiviral defenses triggered by unmethylated CpG, which is recognized as a pathogenic signature by the host (Wang et al., 2016). To evade the host’s immune responses, ASFV may reduce its CpG expression (Tyagi et al., 2017; Mordstein et al., 2021). Another potential reason is the high stack energy of CpG, which could impede translation and replication in DNA duplexes. The lack of overrepresentation of TpG and CpA dinucleotides suggests that methylation and subsequent deamination of cytosine to thymidine is not a major factor, consistent with several studies (Shackelton et al., 2006; Cheng et al., 2013). Interestingly, the slight overrepresentation of TpA similar to the Nipah virus was contrary to many organisms that avoid TpA-containing codons to prevent nonsense mutations (Giallonardo et al., 2017; Khandia et al., 2019; Mordstein et al., 2021).
The constructed phylogenetic tree illustrates the evolutionary relationships between lineages and how natural selection, influenced by geographical factors, shapes ASFV evolution. Some strains nested in foreign clades suggest not only the influence of natural selection but also the potential impact of mutational pressure at the codon level, leading to genotypically similar clades. This inference is supported when a genotype-based tree is constructed; wherein these nested strains no longer appear, reflecting their genotypic relationship with the foreign clades.
Conclusion
African Swine Fever (ASF) is a devastating disease that severely impacts porcine populations, with substantial socio-economic consequences. Therefore, studying the evolutionary biology of the ASF virus (ASFV) is crucial for understanding its replication cycle, complex transmission chain, and adaptation mechanisms. Such understanding can also significantly aid in the development of effective vaccines and therapeutics.
In this study, we conducted a comprehensive exploration of the codon usage bias (CUB) and viral adaptation of ASFV. Our analysis revealed a delicate balance between two major evolutionary forces: natural selection and mutation pressure, which mutually influence the evolution of ASFV. While the viral lineages were distinct and primarily continent-based, they showed a degree of interconnectedness.
Although both natural selection and mutational pressure were found to contribute to ASFV evolution, natural selection appeared to be slightly more predominant. This may be due to the fact that while all viruses experience rapid mutation, ASFV, being a large DNA virus, may not mutate as quickly as RNA viruses. Future live experiments and evolutionary studies are recommended to further investigate this hypothesis.
To our knowledge, this is the first comprehensive analysis of codon usage for the entire ASFV genomic sequences. This work could be valuable for studies investigating viral gene expression and regulation, gene function prediction, parasite-host interaction, and immune dysfunction, and for the design of drugs and vaccines.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
Research idea and design: MK, MC, XC, CW; Resource mobilization and funding: XC, CW; Sample collection and curation: MK, MC, JL, ML; Data analysis, collection and interpretation: MK, MC, XX, YS, JW, YL; Writing and editing: MK, MC, CS, YZ, GY; Supervision and knowledge contribution: XC and CW. The article’s contribution was reviewed and approved by all authors prior to submission.
Funding
This work was supported by the National Natural Science Foundation of China (U21A20261, 32273043, 32202890, 31941018), the Science and Technology Development Program of Jilin Province (20200402041NC), the Science and Technology Development Program of Changchun City (21ZY42) and China Agriculture Research System of MOF and MARA (CARS-35).
Acknowledgments
The authors acknowledge the Chinese Scholarship Council (CSC) for the scholarship facilitation of a PhD student who is among the primary authors of this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2023.11562/full#supplementary-materialhttps://www.frontierspartnerships.org/articles/10.3389/av.2023.11562/full#supplementary-material
SUPPLEMENTARY FIGURE S1Codons most influential in PC1, PC2, PC3, PC1 and PC2, PC2 and PC3, and PC3 and PC4. The first 10 codons that contributed the most are listed on the X-axis, and the Y-axis displays the percentage contribution. TGT, TGC, GAA, GAG, CGC, CAT, CAC, AGA, CTG, and GCG are the codons identified for PC1. TCC, CTC, ACG, GCC, TAT, TAC, ACA, GCA, ATC, and TCG are the codons identified for PC2. CTA, ACC, GTG, CCA, TTG, GAC, GAT, TCA, TTC, and TTT are the codons identified for PC3. Codons TGT, TGC, CGC, GAA, GAG, CTC, TCC, ACG, CTG, and TTA have been identified for the PC1 and PC2 combined model. Codons TCC, CTA, ACC, CTC, GTG, ACG, ACA, GCC, CGT, and GCA have been identified for the PC2 and PC3 combined model. Codons CAC, GAT, CTA, ACC, TTG, ACT, CCA, CGA, CCT, and CTT have been identified for the PC3 and PC4 combined model.
SUPPLEMENTARY TABLE 1Data indicating isolates/strains and ORFs. Isolate information is provided on Sheet 1 and ORF information is provided on Sheet 2. Sheet 1 has six columns; including one for the accession numbers of the isolates, one for the type of isolates, one for the genotypes of isolates, one for the country of origin of the isolates, one for the year of isolation, and one for the Principal Component Analysis (PCA) Identity Number (ID) for ease of reading as indicated on the identified factor map.
SUPPLEMENTARY TABLE 2Proportions of nucleotides and codons at different positions. %A, %C, %T and %G indicate proportion of the overall nucleotide composition of Adenine, Cytosine, Thiamine and Guanine respectively whereas %A3, %C3, %T3 and %G3 indicate proportion of the respective nucleotides at third nucleotide position. %GC indicate overall proportion of GC content whereas %GC3 indicate proportion of GC content at third codon position and %GC12 indicate proportion of GC content at first and second codon position. Effective Number of Codons (ENCs) has been indicated to appreciate the absolute codon usage bias at strain/isolate level.
SUPPLEMENTARY TABLE 3Autocorrelation illustrating correlation between variables. Positive and negative correlations are depicted in numbers with continuous values: p <0.01 indicates very significant correlation while 0.01 < p < 0.05 indicates significant correlation. Zero values indicate absence of correlation.
Abbreviations
ASFV, African Swine Fever Virus; CAI, Codon Adaptation Index; CSC, Chinese Scholarship Council; CUB, Codon Usage Bias; DNA, Deoxyribose Nucleic Acid; ENC, Effective Number of Codons; GRAVY, Grand Average of Hydropathy; IQR, Interquartile Range; MAFFT, Multiple Alignment using Fast Fourier Transform; MAS, Multiple Sequence Alignment; MEGA, Molecular Evolutionary Genetics Analysis; ML, Maximum Likelihood; NWK, Newick; ORF, Opening Reading Frame; PCA, Principal Component Analysis; PR2, Chargaff’s second parity rule; R, Range; RCDI, Relative Codon Deoptimization Index; RNA, Ribose Nucleic Acid; RSCU, Relative Synonymous Codon Usage; SiD, Similarity Index.
References
1
Alkhamis M. A. Gallardo C. Jurado C. Soler A. Sa M. Arias M. (2018). Phylodynamics and evolutionary epidemiology of African swine fever p72-CVR genes in Eurasia and Africa. PLoS ONE13 (2), 01925655–e192618. 10.1371/journal.pone.0192565
2
Andrea L. D. Pintó R. M. Bosch A. Musto H. Cristina J. (2011). A detailed comparative analysis on the overall codon usage patterns in hepatitis a virus. Virus Res.157, 19–24. 10.1016/j.virusres.2011.01.012
3
Anwar A. M. Soudy M. Mohamed R. (2020). “vhcub: virus-host codon usage co-adaptation analysis [version 1; peer review:2 approved],” in F1000Res.8 (2137), 1–10. Available at: https://doi.org/org/10.12688/f1000research.21763.1.
4
Ata E. B. Li Z. J. Shi C. W. Yang G. L. Yang W. T. Wang C. F. (2022). African swine fever virus: a raised global upsurge and a continuous threaten to pig husbandry. Microb. Pathog.167, 105561. 10.1016/J.MICPATH.2022.105561
5
Ata G. Wang H. Bai H. Yao X. Tao S. (2021). Edging on mutational bias, induced natural selection from host and natural reservoirs predominates codon usage evolution in hantaan virus. Front. Microbiol.12, 699788–699818. 10.3389/fmicb.2021.699788
6
Ayanwale A. Trapp S. Guabiraba R. Caballero I. Roesch F. (2022). New insights in the interplay between african swine fever virus and innate immunity and its impact on viral pathogenicity. Front. Microbiol.13, 958307–958309. 10.3389/fmicb.2022.958307
7
Bera B. C. Virmani N. Kumar N. Anand T. Pavulraj S. Rash A. et al (2017). Genetic and codon usage bias analyses of polymerase genes of equine influenza virus and its relation to evolution. BMC Genomics18, 652–718. 10.1186/s12864-017-4063-1
8
Brown V. R. Bevins S. N. (2018). A review of african swine fever and the potential for introduction into the United States and the possibility of subsequent establishment in feral swine and native ticks. Front. Veterinary Sci.5, 11–18. 10.3389/fvets.2018.00011
9
Butt A. M. Nasrullah I. Qamar R. Tong Y. (2016). Evolution of codon usage in zika virus genomes is host and vector specific. Emerg. Microbes Infect.5, 1–14. 10.1038/emi.2016.106
10
Butt A. M. Nasrullah I. Tong Y. (2014). Genome-wide analysis of codon usage and influencing factors in chikungunya viruses. PLoS ONE9 (3), e90905–e90920. 10.1371/journal.pone.0090905
11
Chen S. N. Li C. L. Lin J. S. Zhai S. L. Sun M. F. (2022). Diverse african swine fever viruses in China. New Microbes New Infect.46, 100976. 10.1016/j.nmni.2022.100976
12
Chen Y. Shi Y. Deng H. Gu T. Xu J. Ou J. et al (2014). Characterization of the porcine epidemic diarrhea virus codon usage bias. Infect. Genet. Evol.28, 95–100. 10.1016/j.meegid.2014.09.004
13
Chen Y. Xu Q. Yuan X. Li X. Zhu T. Ma Y. et al (2017). Analysis of the codon usage pattern in middle East respiratory syndrome coronavirus. Oncotarget8 (66), 110337–110349. 10.18632/oncotarget.22738
14
Cheng X. Virk N. Chen W. Ji S. Ji S. Sun Y. et al (2013). CpG usage in RNA viruses: data and hypotheses. PLoS ONE8 (9), e74109–9. 10.1371/journal.pone.0074109
15
Comeron J. M. Aguade M. (1998). An evaluation of measures of synonymous codon usage bias. J. Mol. Evol.47, 268–274. 10.1007/pl00006384
16
Costard S. Wieland B. Glanville W. D. Jori F. Rowlands R. Vosloo W. et al (2009). African swine fever: how can global spread be prevented. Philosophical Trans. R. Soc. B364, 2683–2696. 10.1098/rstb.2009.0098
17
Craig A. F. Schade-Weskott M. L. Harris H. J. Heath L. Kriel G. J. P. Schalkwyk L. V. et al (2021). Extension of sylvatic circulation of african swine fever virus in extralimital warthogs in South Africa. Front. Veterinary Sci.8, 746129–746211. 10.3389/fvets.2021.746129
18
Deb B. Uddin A. Chakraborty S. (2020). Codon usage pattern and its influencing factors in different genomes of hepadnaviruses. Archives Virology165, 557–570. 10.1007/s00705-020-04533-6
19
Deb B. Uddin A. Chakraborty S. (2021a). Composition, codon usage pattern, protein properties, and influencing factors in the genomes of members of the family anelloviridae. Archives Virology166 (0123456789), 461–474. 10.1007/s00705-020-04890-2
20
Deb B. Uddin A. Chakraborty S. (2021b). Genome-wide analysis of codon usage pattern in herpesviruses and its relation to evolution. Virus Res.292, 198248. 10.1016/j.virusres.2020.198248
21
Dixon L. K. Chapman D. A. G. Netherton C. L. Upton C. (2012). African swine fever virus replication and genomics. Virus Res.173 (1), 3–14. 10.1016/j.virusres.2012.10.020
22
Giallonardo F. D. Schlub T. E. Shi M. Holmes E. C. (2017). Dinucleotide composition in animal RNA viruses is shaped more by virus family than by host species. J. Virology91 (8), 1–15. 10.1128/JVI.02381-16
23
Gu H. Chu D. K. W. Peiris M. Poon L. L. M. (2020). Multivariate analyses of codon usage of SARS-CoV-2 and other betacoronaviruses. Virus Evol.6 (1), veaa032–10. 10.1093/ve/veaa032
24
He W. Wang N. Tan J. Wang R. Yang Y. Li G. et al (2019). Comprehensive codon usage analysis of porcine deltacoronavirus. Mol. Phylogenetics Evol.141, 106618–106710. 10.1016/j.ympev.2019.106618
25
Hemert F. V. van der Kuyl A. C. Berkhout B. (2016). Impact of the biased nucleotide composition of viral RNA genomes on RNA structure and codon usage. J. General Virology97, 2608–2619. 10.1099/jgv.0.000579
26
Hou W. (2020). Characterization of codon usage pattern in SARS-CoV-2. Virology J.17, 138–210. 10.1186/s12985-020-01395-x
27
Husson F. Josse J. Le S. Mazet J. (2022). FactoMineR: multivariate exploratory data analysis and data mining, the comprehensive R archive network. Available at: https://cran.r-project.org/package=FactoMineR (Accessed December 20, 2022).
28
Iyer L. M. Aravind L. Koonin E. V. (2001). Common origin of four diverse families of large eukaryotic DNA viruses. J. Virology75 (23), 11720–11734. 10.1128/jvi.75.23.11720-11734.2001
29
Jia N. Ou Y. Pejsak Z. Zhang Y. Zhang J. (2017). Roles of African swine fever virus structural proteins in viral infection. J. Veterinary Res.61, 135–143. 10.1515/jvetres-2017-0017
30
Karlin S. Doerfler W. Cardon L. R. (1994). Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses?J. Virology68 (5), 2889–2897. 10.1128/jvi.68.5.2889-2897.1994
31
Kassambara A. Mundt F. (2022). factoextra: extract and visualize the results of multivariate data analyses, the comprehensive R archive network. Available at: https://cran.r-project.org/package=factoextra (Accessed December 10, 2022).
32
Khandia R. Singhal S. Kumar U. Ansari A. Tiwari R. Dhama K. et al (2019). Analysis of Nipah virus codon usage and adaptation to hosts. Front. Microbiol.10, 886–918. 10.3389/fmicb.2019.00886
33
Kumar N. Kaushik R. Tennakoon C. Uversky V. N. Mishra A. Sood R. et al (2021). Evolutionary signatures governing the codon usage bias in coronaviruses and their implications for viruses infecting various bat species. Viruses13, 1847–1918. 10.3390/v13091847
34
Kumar N. Kulkarni D. D. Lee B. Kaushik R. Bhatia S. Sood R. et al (2018). Evolution of codon usage bias in henipaviruses is governed by natural selection and is host-specific. Viruses10, 604. 10.3390/v10110604
35
Lobry J. Gautier C. (1994). Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes. Nucleic Acids Res.22 (15), 3174–3180. 10.1093/nar/22.15.3174
36
Loh J. Zhao G. Presti R. M. Holtz L. R. Finkbeiner S. R. Droit L. et al (2009). Detection of novel sequences related to african swine fever virus in human serum and sewage. J. Virology83 (24), 13019–13025. 10.1128/JVI.00638-09
37
Lubisi B. A. Bastos A. D. S. Dwarka R. M. Vosloo W. (2005). Molecular epidemiology of african swine fever in East Africa. Archives Virology150, 2439–2452. 10.1007/s00705-005-0602-1
38
Ma X. Chang Q. Ma P. Li L. Zhou X. Zhang D. et al (2017). Analyses of nucleotide, codon and amino acids usages between peste des petits ruminants virus and rinderpest virus. Gene637, 115–123. 10.1016/j.gene.2017.09.045
39
Mordstein C. Cano L. Morales A. C. Young B. Ho A. T. Rice A. M. et al (2021). Transcription, mRNA export, and immune evasion shape the codon usage of viruses. Genome Biol. Evol.13 (9), evab106–14. 10.1093/gbe/evab106
40
Mueller S. Papamichail D. Coleman J. R. Skiena S. Wimmer E. (2006). Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virology80 (19), 9687–9696. 10.1128/JVI.00738-06
41
Nakamura Y. Gojobori T. Ikemura T. (2000). Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res.28 (1), 292. 10.1093/nar/28.1.292
42
Nasrullah I. Butt A. M. Tahir S. Idrees M. Tong Y. (2015). Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on marburg virus evolution. BMC Evol. Biol.15, 174–215. 10.1186/s12862-015-0456-4
43
Peng Q. Ni Y. Zhang X. Liu M. Li J. Li B. et al (2022). Comprehensive analysis of codon usage patterns of porcine deltacoronavirus and its host adaptability. Transbound. Emerg. Dis.2022, e2443–e2455. 10.1111/tbed.14588
44
Pu F. Wang R. Yang X. Hu X. Wang J. Zhang L. et al (2023). Nucleotide and codon usage biases involved in the evolution of african swine fever virus: a comparative genomics analysis. J. Basic Microbiol.2023, 499–518. 10.1002/jobm.202200624
45
Puigbò P. Aragonès L. Garcia-vallvé S. (2010). RCDI/eRCDI: a web-server to estimate codon usage deoptimization. BMC Res. Notes3, 87–95. 10.1186/1756-0500-3-87
46
Puigbò P. Bravo I. G. Garcia-vallve S. (2008). CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct8, 38–8. 10.1186/1745-6150-3-38
47
Salguero F. J. (2020). Comparative pathology and pathogenesis of african swine fever infection in swine. Front. Veterinary Sci.7, 282–314. 10.3389/fvets.2020.00282
48
Sang H. Miller G. Lokhandwala S. Sangewar N. Waghela S. D. Bishop R. P. et al (2020). Progress toward development of effective and safe african swine fever virus vaccines. Front. Veterinary Sci.7, 84–89. 10.3389/fvets.2020.00084
49
Shackelton L. A. Parrish C. R. Holmes E. C. (2006). Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J. Mol. Evol.62, 551–563. 10.1007/s00239-005-0221-1
50
Sharp P. M. Li W.-H. (1986). An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol.24, 28–38. 10.1007/bf02099948
51
Sharp P. M. Li W. (1987). The codon adaptation index - a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res.15 (3), 1281–1295. 10.1093/nar/15.3.1281
52
Shi S.-L. Jiang Y.-R. Liu Y.-Q. Xia R.-X. Qin L. (2013). Selective pressure dominates the synonymous codon usage in parvoviridae. Virus Genes46, 10–19. 10.1007/s11262-012-0818-6
53
Sueoka N. (1988). Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci.85, 2653–2657. 10.1073/pnas.85.8.2653
54
Sueoka N. (1999a). Translation-coupled violation of parity Rule 2 in human genes is not the cause of heterogeneity of the DNA G + C content of third codon position. Gene238, 53–58. 10.1016/s0378-1119(99)00320-0
55
Sueoka N. (1999b). Two aspects of DNA base composition: G + C content and translation-coupled deviation from intra-strand rule of A = T and G = C. J. Mol. Evol.49, 49–62. 10.1007/pl00006534
56
Sun J. Zhao W. Wang R. Zhang W. Li G. Lu M. et al (2020). Analysis of the codon usage pattern of ha and na genes of H7N9 influenza A virus. Int. J. Mol. Sci.21, 7129–7221. 10.3390/ijms21197129
57
Tao P. Dai L. Luo M. Tang F. Tien P. Pan Z. (2009). Analysis of synonymous codon usage in classical swine fever virus. Virus Genes38, 104–112. 10.1007/s11262-008-0296-z
58
Teklue T. Wang T. Luo Y. Hu R. Sun Y. Qiu H. (2020). Generation and evaluation of an african swine fever virus mutant with deletion of the CD2v and UK genes. Vaccines8, 763–817. 10.3390/vaccines8040763
59
Tian L. Shen X. Murphy R. W. Shen Y. (2018). The adaptation of codon usage of + ssRNA viruses to their hosts. Infect. Genet. Evol.63, 175–179. 10.1016/j.meegid.2018.05.034
60
Todorov H. Fournier D. Gerber S. (2018). Principal components analysis: theory and application to gene expression data analysis. Genomics Comput. Biol.4 (2), 100041–100111. 10.18547/gcb.2018.vol4.iss2.e100041
61
Tyagi A. Kumar B. T. N. Singh N. K. (2017). Genome dynamics and evolution of codon usage patterns in shrimp viruses. Archives Virology162 (10), 3137–3142. 10.1007/s00705-017-3445-7
62
Urbano A. C. Ferreira F. (2020). Role of the dna-binding protein pa104r in asfv genome packaging and as a novel target for vaccine and drug development. Vaccines8 (4), 585–617. 10.3390/vaccines8040585
63
Wang G. Xie M. Wu W. Chen Z. (2021). Structures and functional diversities of asfv proteins. Viruses13 (11), 2124. 10.3390/v13112124
64
Wang H. Liu S. Zhang B. Wei W. (2016). Analysis of synonymous codon usage bias of zika virus and its adaption to the hosts. PLoS ONE11 (11), 01662600–e166322. 10.1371/journal.pone.0166260
65
Wickham H. Chang W. Henry L. Pedersen T. L Takahashi K. Wilke C. et al (2022). ggplot2: create elegant data visualisations using the grammar of graphics, the comprehensive R archive network. Available at: https://cran.r-project.org/package=ggplot2 (Accessed November 10, 2022).
66
Wold S. Esbensen K. Geladi P. (1987). Principal component analysis. Chemom. Intelligent Laboratory Syst.2, 37–52. 10.1016/0169-7439(87)80084-9
67
Wright F. (1990). The “effective number of codons” used in a gene. Gene87, 23–29. 10.1016/0378-1119(90)90491-9
68
Yao H. Chen M. Tang Z. (2019). Analysis of synonymous codon usage bias in flaviviridae virus. BioMed Res. Int.2019, 1–12. 10.1155/2019/5857285
69
Yao X. Fan Q. Yao B. Lu P. Rahman S. U. Chen D. et al (2020). Codon usage bias analysis of bluetongue virus causing livestock infection. Front. Microbiol.11, 655–712. 10.3389/fmicb.2020.00655
70
Yu X. Liu J. Li H. Liu B. Zhao B. Ning Z. (2021a). Comprehensive analysis of synonymous codon usage bias for complete genomes and E2 gene of atypical porcine pestivirus. Biochem. Genet.59, 799–812. 10.1007/s10528-021-10037-y
71
Yu X. Liu J. Li H. Liu B. Zhao B. Ning Z. (2021b). Comprehensive analysis of synonymous codon usage patterns and influencing factors of porcine epidemic diarrhea virus. Archives Virology166, 157–165. 10.1007/s00705-020-04857-3
72
Zhang J. Wang M. Liu W. Zhou J. Chen H. Ma L. et al (2011). Analysis of codon usage and nucleotide composition bias in polioviruses. Virology J.8, 146–148. 10.1186/1743-422X-8-146
73
Zhang K. Yang B. Shen C. Zhang T. Hao Y. Zhang D. et al (2022). MGF360-9L is a major virulence factor associated with the african swine fever virus by antagonizing the JAK/STAT signaling pathway. mBio13 (1), 1–19. 10.1128/MBIO.02330-21
74
Zhang X. Cai Y. Zhai X. Liu J. Zhao W. Ji S. et al (2018). Comprehensive analysis of codon usage on rabies virus and other lyssaviruses. Int. J. Mol. Sci.19, 2397–2415. 10.3390/ijms19082397
75
Zheng X. Nie S. Feng W. (2022). Regulation of antiviral immune response by African swine fever virus (ASFV). Virol. Sin.37 (2), 157–167. 10.1016/j.virs.2022.03.006
76
Zhou J. Gao Z. Sun D. Ding Y. Zhang J. Stipkovits L. et al (2013). A comparative analysis on the synonymous codon usage pattern in viral functional genes and their translational initiation region of ASFV. Virus Genes46, 271–279. 10.1007/s11262-012-0847-1
77
Zhou J. Zhang J. Sun D. Ma Q. Chen H. Ma L. et al (2013). The distribution of synonymous codon choice in the translation initiation region of dengue virus. PLoS ONE8 (10), 772399–e77247. 10.1371/journal.pone.0077239
Summary
Keywords
ASFV, codon usage bias, whole genome, continental lineages, phylogenic tree
Citation
Kanyema MM, Cheng M, Luo J, Lu M, Xing X, Sun Y, Wang J, Lu Y, Shi C, Zeng Y, Yang G, Cao X and Wang C (2023) Comprehensive codon usage analysis of the African Swine Fever Virus. Acta Virol. 67:11562. doi: 10.3389/av.2023.11562
Received
10 May 2023
Accepted
25 August 2023
Published
18 September 2023
Volume
67 - 2023
Edited by
Boris Klempa, Slovak Academy of Sciences, Slovakia
Reviewed by
Kanisht Batra, Lala Lajpat Rai University of Veterinary and Animal Sciences, India
Niraj Kumar Singh, Bihar Animal Sciences University, India
Updates
Copyright
© 2023 Kanyema, Cheng, Luo, Lu, Xing, Sun, Wang, Lu, Shi, Zeng, Yang, Cao and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Cao, xinc@jlau.edu.cn; Chunfeng Wang, wangchunfeng@jlau.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.