Abstract
TRK-fused gene (TFG, tropomyosin-receptor kinase fused gene) is known to negatively regulate the retinoic acid inducible gene (RIG)-I-like receptor (RLR)-mediated interferon (IFN)-I pathway in human cells, thereby participating in the paramyxovirus infection process. We showed that pigeon paramyxovirus type 1 (PPMV-1) infection significantly upregulates TFG expression in infected cells at an early stage. We speculated that PPMV-1 would inhibit IFN activation by upregulating a negative regulator of the IFN pathway. This hypothesis was proved when TFG protein expression was knocked down by RNAi and the replication level of PPMV-1 virus decreased, which indicated that TFG upregulation in the early infection stage benefit virus replication. We next used the IFN-β promoter reporter system to evaluate the role of the TFG in the IFN pathway. The results showed that the TFG inhibited the IFN-β expression stimulated by RIG-I, MAVS (mitochondrial antiviral signaling protein) and TANK-binding kinase 1 (TBK1), but did not inhibit IFN-β activated by the interferon regulatory transcription factor 3 (IRF3), indicating that TFG may affect the function of TBK1, which play an important role in phosphorylation of the IRF3. Further experiments showed that the TFG inhibited the phosphorylation of TBK1, resulting in IRF3 being unable to be phosphorylated. Subsequent experiments on IFN pathway activation confirmed that the IRF3 phosphorylation level was significantly downregulated after overexpression of TFG, while the IFN-β promoter reporting experiment showed that TFG did not directly inhibit the IFN response activated by IRF3. This confirmed that TFG protein negatively regulates the IFN-β pathway by inhibiting TBK1 phosphorylation.
Introduction
The Paramyxoviridae family contains single-stranded, negative-sense enveloped RNA viruses that usually infect vertebrates including mammals, reptiles, birds, and fish (Rima et al., 2019). Almost all paramyxoviruses cause acute respiratory diseases and several members are extremely contagious (Parks and Alexander-Miller, 2013). Among them, Mumps virus, Measles virus, Respiratory Syncytial virus, Parainfluenza virus, Nipah virus, and Hendra virus cause human diseases (Zeltina et al., 2016; Thibault et al., 2017). While Sendai and Canine Distemper viruses cause respiratory manifestations in mice and canines, respectively (Faísca and Desmecht, 2007; Sawatsky et al., 2018), avian Metapneumovirus and avian Paramyxovirus (APMV) cause severe economic losses worldwide in wild and domestic avian species (Ganar et al., 2014; Gogoi et al., 2017). APMV serotype-1 (APMV-1), which is also known as Newcastle disease virus, was renamed “avian orthoavulavirus 1” in the latest update from the International Committee on Taxonomy of Viruses.1 APMV-1 infections cause strong inflammatory responses in their hosts. These responses are triggered by viral replication in infected cells or tissues, leading to excessive cell apoptosis, tissue damage, and significantly increased cytokine/chemokine production (Hu et al., 2012; Meng et al., 2012; Rajmani et al., 2015; Ginting et al., 2017; Zhao et al., 2019; Zhang et al., 2023).
Pigeon paramyxovirus type 1 (PPMV-1), an antigenic variant of APMV-1 that circulates in pigeons, can infect poultry and wild birds (Kommers et al., 2001; Guo et al., 2014). The first report of PPMV-1 was in the late 1970s in Middle Eastern countries (Kaleta et al., 1985). PPMV-1 was first identified in 1985 in Hong Kong (Liu et al., 2006). PPMV-1 is a most dangerous pathogen for doves and pigeons in China (Guo et al., 2013; Wang et al., 2017; He et al., 2020). The PPMV-1 currently circulating in China belongs to genotype VI (Tian et al., 2020; Yu et al., 2022; Zhan et al., 2022). The PPMV-1 SD19 strain used in the present study was isolated in Shandong province from an outbreak in a pigeon yard. Although most of the PPMV-1 isolates are non-pathogenic to chickens, they can cause paralysis, torticollis, diarrhea, and high morbidity, and even sudden death in pigeons (Śmietanka et al., 2014). New evidence indicates that the pathogenicity of PPMV-1 may related to the age of a pigeon (Xie et al., 2020). There is no commercial vaccine available for Newcastle Disease (ND) prevention in pigeons in China. APMV-1 genotype II vaccines for chickens, such as La Sota, have been used to protect pigeons from ND. However, the protective efficacy of these vaccines is not ideal (Liu et al., 2015).
The innate immune system, including interferon production and natural killer cell activation, plays a critical role in the antiviral response. Innate immune system activation requires the recognition of pathogen-associated molecular patterns on invading viruses by pattern-recognition receptors (PRRs) (Akira, 2009). The most important PRRs for RNA viruses are retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), both of which contain DExD/H-box RNA helicase domains that can directly detect viral RNA (Yoneyama et al., 2004). After identifying viral RNA, two caspase recruitment domains (CARDs) at the N-terminal of RIG-I and MDA5 interact with the CARD domain of the mitochondrial antiviral signaling protein (MAVS) to initiate downstream signaling (Seth et al., 2005). The interaction between RIG-I/MDA5 and MAVS activates the TANK-binding kinase 1 (TBK1) signal, leading to phosphorylation of TBK1 (Hou et al., 2011). Phosphorylated TBK1 activates downstream IRF3 phosphorylation, and the resulting phosphorylated IRF3 protein-formed dimer is transferred to the cell nucleus to initiate IFN transcription from where it activates downstream antiviral immunity (Lee et al., 2009).
The TFG, a evolutionarily conserved secretion system regulator protein, is wrapped around COPII (coat protein complex II) vesicles in the form of polymers, which help the vesicles budding from the endoplasmic reticulum effectively from the endoplasmic reticulum and fusing with the Golgi intermediate from the endoplasmic reticulum (Johnson et al., 2015; Hanna et al., 2017; Peotter et al., 2019). In addition to its role in intracellular transport, TFG in pancreatic β-cells maintains the mass and function of β-cells, and its dysfunction is associated with glucose intolerance (Yamamotoya et al., 2017). In recent years, cumulative evidence has shown that TFG is associated with the immune response. Miranda et al. (2006) found that TFG is a NF-κB (Nuclear factor-κB) activator and interacts with IKK-γ (nuclear factor kappa-B kinase subunit gamma) and TANK. Wynne et al. (2014) reported that tripartite motif (TRIM) 68 can increase TFG degradation and turn off TFG-mediated IFN-β production. Interestingly, Lee et al. (2013a) found that TFG protein acts as a negative regulator of the IFN pathway, and inhibits the IFN response activated by RIG-I. However, the molecular mechanism by which TFG inhibits RIG-I-mediated IFN pathway activation is still unclear. In the present study, after infecting host cells with PPMV-1, the expression level of TFG was significantly upregulated in the early stage of infection. Thus, we further investigated the role of TFG protein in PPMV-1 virus replication and the molecular mechanism underlying TFG protein inhibition of the IFN pathway.
Materials and methods
Cells and viruses
The human cervical cancer cell line (HeLa) and chicken embryo fibroblast monolayer cell line (DF-1) were purchased from ATCC (Manassas, VA, United States). These cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermo, Grand Island, NY, United States) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco), 1% Penicillin-Streptomycin (×100, Macgene, Beijing, China) and maintained in DMEM containing 2% FBS. The cells were kept in a 5% CO2 incubator at 37°C (Thermo Forma, Hamilton, NJ, United States). The genotype VI PPMV-1 strain SD19 was isolated in Shandong Province from an outbreak in a pigeon yard as previously reported (Tian et al., 2020).
Reagents
Anti-TFG (HPA052206) and Flag (M2 F-1804) antibodies were purchased from Sigma (St. Louis, MO). Antibodies against β-actin (4970), TBK1 (3504), Phospho-TBK1 (Ser172) (5483), IRF-3 (4302), and Phospho-IRF3 (Ser396) (29047) were purchased from Cell Signaling Technology. The antibody against PPMV-1 nucleocapsid protein (NP) was prepared by our laboratory. Poly (I:C) were purchased from Sigma-Aldrich (St. Louis, MO).
Plasmids and transfection
Flag-tagged RIG-I, MDA-5, MAVS, TBK-1, IKK-ε and IRF3 were provided by C. Ding (Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Science, Shanghai, China). TFG cDNAs were amplified by PCR using cDNAs made from Hela cells as the template and then cloned into Flag-N1 vector. The non-structural protein 1 (NS1) fragment of influenza virus H1N1 was amplified by PCR using cDNAs from H1N1-infected Hela cells as the template and then cloned into p3XFLAG-CMV-14 (Sigma) vector. All the constructs were confirmed by DNA sequencing. A list of PCR primer is provided in Table 1. For reporter assays, the IFN-β promoter luciferase reporter (p-125Luc) and the IFN sequence response element (ISRE) luciferase reporters (pISRE-TA-luc) were provided by C. Ding (Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Science, Shanghai, China). Plasmids were transfected into Hela cells using Lipo2000 transfection reagent (Thermo Fisher) according to the manufacturer’s protocol.
TABLE 1
| Primer (restriction enzyme site) | Sequence (5′-3′) |
|---|---|
| TFG-F (XhoI) | CCGCTCGAGATGAACGGACAGTTGGATCTAAGTG |
| TFG-R (BamHI) | CGCGGATCCCGATAACCAGGTCCAGGTTGG |
| NS1-F (XhoI) | CGCTCGAGATGGACTCCAACACCATGTCAAGCTT |
| NS1-R (NotI) | GGTGGCGGCCGCTCATTTCTGCTCTGGAGGTAGTGAA |
Primers used for plasmids construction.
Quantitative reverse-transcriptase PCR (qRT-PCR)
Total RNA was isolated from virus-infected HeLa cells or normal HeLa cells using the Total RNA Isolation Kit from Foregene (Chengdu, China) according to the manufacturer’s protocol. Total RNA (1 μg/sample) was reverse transcribed into cDNA using PrimeScript RT Master MIX (Takara Biomedical Technology, Beijing, China). qRT-PCR was performed using SYBR® Premix Ex Taq™ Kit (Takara). Each reaction was performed in a 20 µL volume containing 8 µL of 2× SYBR® Premix Ex Taq™, 1 µL (10 µg) of each primer, and 10 µL cDNA. The cycling parameters were 1 cycle at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, and 60°C for 30 s. One additional cycle was added for the melting curve analysis with all reactions to verify product specificity. The relative gene expression levels were normalized to that of the human 18s gene. The threshold cycle 2−△△CT method was used to determine the fold change of gene expression levels. TFG was PCR-amplified using the primer pair human-TFG-RT-F 5′-GAACGGACAGTTG-3′ and human-TFG-RT-R 5′-CCGATAACCA GGT-3′. TFG mRNA levels were normalized to those of the human 18s gene, which involved PCR amplification with the primer pair human-18S-RT-F 5′-CGGCTACCACATCCAAGGAA-3′ and human-18S-RT-R 5′-GCTGGAATTACCGCGGCT-3′.
Western blotting
Whole-cell lysates were prepared in RIPA buffer (Beyotime Biotechnology, Shanghai, China) for total protein extraction. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer was added to 30 µg of each cell lysate and the lysates were resolved by SDS-PAGE and then transferred electrophoretically onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% skimmed milk powder in PBST buffer (PBS containing 0.5% Tween 20) for 1 h at 37°C and then incubated overnight at 4°C with the corresponding primary antibodies diluted 1:1,000. The membranes were washed three times with PBST buffer and incubated for 1 h at room temperature (RT) with corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies diluted 1:10,000. HRP was detected with Western Lightning Chemiluminescence Reagent (CWBIO, Beijing, China). The relative levels of the selected proteins to actin expression were determined by densitometry using Image J software version 1.8.0.
Indirect immunofluorescence assay
HeLa cells cultured in 6-well plates were infected with PPMV-1 at a MOI of 0.1 and prepared for indirect immunofluorescence analysis at 24 and 48 h post-infection (hpi). Briefly, cells were collected at the stipulated times and rinsed three times with phosphate-buffered saline (PBS), fixed with 300 µL 4% paraformaldehyde (30 min, RT), and then permeabilized with 300 µL 0.2% Triton X-100 (Sigma) in PBS per well for 10 min. Cells were rinsed three times with PBS and blocked with 200 µL skimmed milk (30 min, RT), and then incubated with 200 µL primary antibody diluted 1:1,000 in skimmed milk (1 h, 37°C). After three PBS washes, the cells were incubated with anti-FITC or anti-TRITC secondary antibodies (Beyotime) (30 min, RT). Cells were rinsed three times with PBS and then counterstained with DAPI (4,6- diamidino-2-phenylindole) diluted 1:1,000 (Sigma-Aldrich, St. Louis, MO, United States) to detect the nuclei. Images were captured with the ImageXpress Micro Confocal High-Content Imaging System (Molecular Devices) and processed with Adobe Photoshop CS5 software.
siRNA treatment and virus infection
siRNA TFG #1 (5′-GUCAGAUUGAA GGUCAGAUTT-3′) and TFG #2 (5′-AUCUGACCUUCAAUCUGACTT-3′) were designed to knockdown TFG expression in HeLa cells. For transfection with the siRNA against TFG, HeLa cells were transfected with 4 µL siRNA using 8 µL Lipofectamine® RNAiMAX (Thermo Fisher) in Opti-MEM medium following the manufacturer’s instructions. The knockdown efficiency was measured by detecting endogenous protein expression by Western blot analysis. To study the effect of TFG on the replication of PPMV-1, strain SD19 (MOI = 0.1) was used to infect TFG-siRNA- or Mock-siRNA-treated HeLa cells at 24 h post transfection. Viral NP protein was detected at 12, 24, 36 and 48 hpi.
Dual luciferase assay
Hela cells were seeded into 24-well plates 12–16 h before transfection. When the cell density reached ∼60%–70%, 0.3 µg of the TFG expression plasmid or its corresponding empty vector was transfected, and each well was transfected with the 0.3 µg of designated IFN pathway stimulation protein, 100 ng ISRE reporter plasmid, and 20 ng sea renal luciferase reporter plasmid (pTK), with pTK as the internal reference. The supernatant was discarded after 24 h, the cells were washed with PBS twice, and 100 µL of the diluted cell lysates were added to each well, followed by plate shaking (15 min, RT). Lysate from each well (30 µL) was analyzed with a dual luciferase assay kit (Promega, Madison, WI, United States) to determine the luciferase activity according to the manufacturer’s instructions. A unit of relative luciferase activity was defined as the ratio of FLuc activity to RLuc activity. Luciferase expression was measured in triplicate in each experiment, and three separate experiments were performed.
Tissue culture infectious dose 50 (TCID50) assay
Virus titers in culture medium of PPMV-1-infected cells was determined by measuring TCID50 in DF-1 cells. In brief, DF-1 cells were seeded in 96-well plates at a density of 2.0 × 104 cells per well. After 24 h, cells were infected with virus, which was serially diluted in 10-fold using serum free medium. The virus and cells were incubated at 37°C for 4 days. The cytopathic effect of cells was observed using light microscopy. TCID50 was calculated by the Karber method.
Enzyme-linked immunosorbent assay (ELISA) for IFN-β
An ELISA to quantify secreted IFN-β was carried out on culture supernatants collected from infected cells. Medium was collected and centrifuged to remove cell debris. Fifty microliters of cleared supernatant or the IFN-β standard was used in duplicate for detection of IFN-β by using a human IFN-β ELISA kit (PBL IFN Source, Piscataway, NJ) according to the manufacturer’s instructions.
Statistical analyses
Statistical analyses were performed using GraphPad Prism software to perform Student’s t-test or analysis of variance (ANOVA) on at least three independent replicates. p values of < 0.05 were considered statistically significant for each test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Results
TFG expression increases in the early stages of PPMV-1 infection in HeLa cells
To detect changes in TFG mRNA level after PPMV-1 infection, HeLa cells were infected with PPMV-1 at a multiplicity of infection (MOI) of 0.1, and TFG mRNA expression was detected at 12, 24, 36 and 48 hpi. TFG mRNA levels significantly increased at 12 hpi, but no significant difference between the infected group and the control group was observed at 24 hpi (Figure 1A). TFG protein expression in PPMV-1 infected cells was also determined. HeLa cells were infected with PPMV-1 at a MOI of 0.01 (Figure 1B), and the cells were harvested at different time points post-infection for Western blotting analysis. The TFG protein expression levels were found to significantly increase at 12 hpi (Figure 1C), whereas the TFG protein levels in the infected group were significantly lower than those in the mock control at 36 and 48 hpi. These data show that the TFG was upregulated in the early stage of PPMV-1 infection.
FIGURE 1
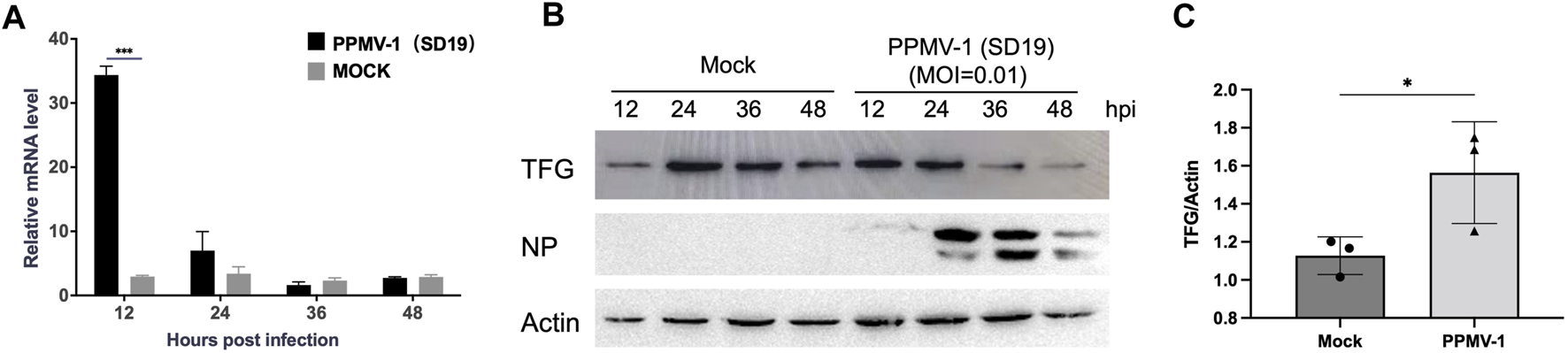
TFG expression increased in the early stage of HeLa cell infection with PPMV-1. (A) TFG mRNA expression levels in HeLa cells infected with PPMV-1 were measured by qRT-PCR. The relative expression levels of TFG were compared with expression of the human 18s gene. (B) TFG protein expression levels in HeLa cells infected with PPMV-1 at MOI of 0.01 were examined by Western blotting. (C) The relative levels of TFG in PPMV-1 infected cells to that in mock cells at 12 hpi were determined by densitometry using Image J software version 1.8.0. The data represent the means ± the standard deviations (SD) of three independent experiments. Statistical analyses were performed using GraphPad Prism software (*, p < 0.05; ***, p < 0.001).
Knocking down TFG inhibited PPMV-1 replication by modulating the IFN pathway
To learn about the intracellular localization of PPMV-1 and TFG in detail, we compared the subcellular localization of PPMV-1-infected HeLa cells at different time points. As shown in Figure 2, TFG protein was distributed diffusely in the cytoplasm. PPMV-1 infection did not affect TFG protein localization. Most of the PPMV-1 NP accumulated around the nucleus at 24 hpi, becoming cytoplasmically localized at 48 hpi. Co-localization of the NP protein and TFG was observed in some infected cells. These data possibly indicate that TFG play a role in PPMV-1 infection. To further investigate the effect of TFG protein on PPMV-1 replication, RNAi was used to knock down TFG expression levels. The expression levels of NP, TFG and actin proteins in PPMV-1-infected HeLa cells were examined by Western blotting, the virus titers in supernatant were determined by TCID50 assay in DF-1 cells. As shown in Figures 3A, B, after knocking down the TFG protein, PPMV-1 NP protein levels were significantly lower than those of the mock control at 36 and 48 hpi (Figure 3A), the virus titers in supernatant were also lower than that of control group at 36 hpi (Figure 3B), indicating that the existence of TFG protein is of benefit to viral replication.
FIGURE 2
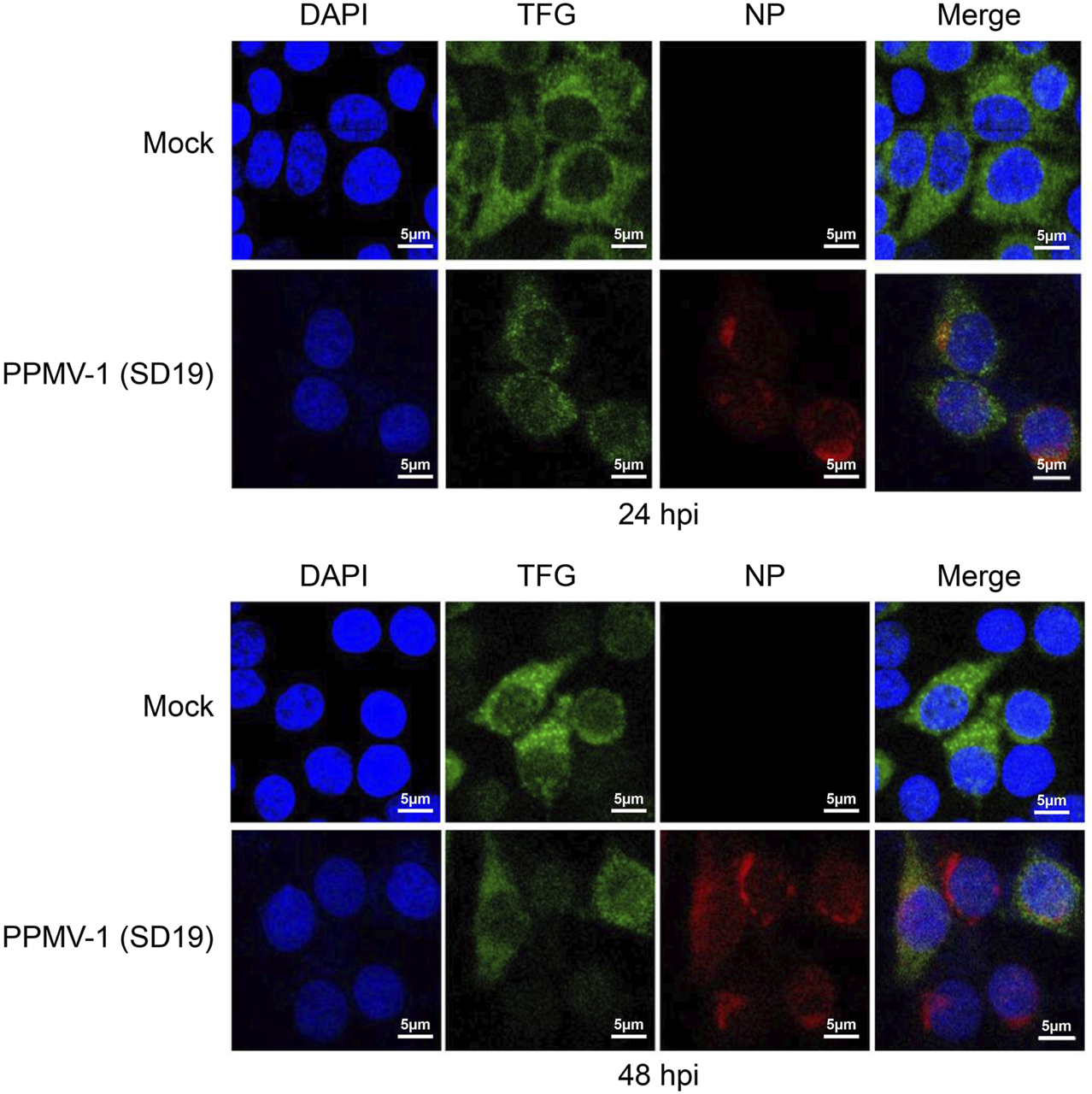
Subcellular localization of PPMV-1 and TFG in HeLa cells. The subcellular localization of TFG protein in PPMV-1-infected HeLa cells at 24 and 48 hpi. DAPI was used to stain the nuclei. Scale bars, 5 μm.
FIGURE 3
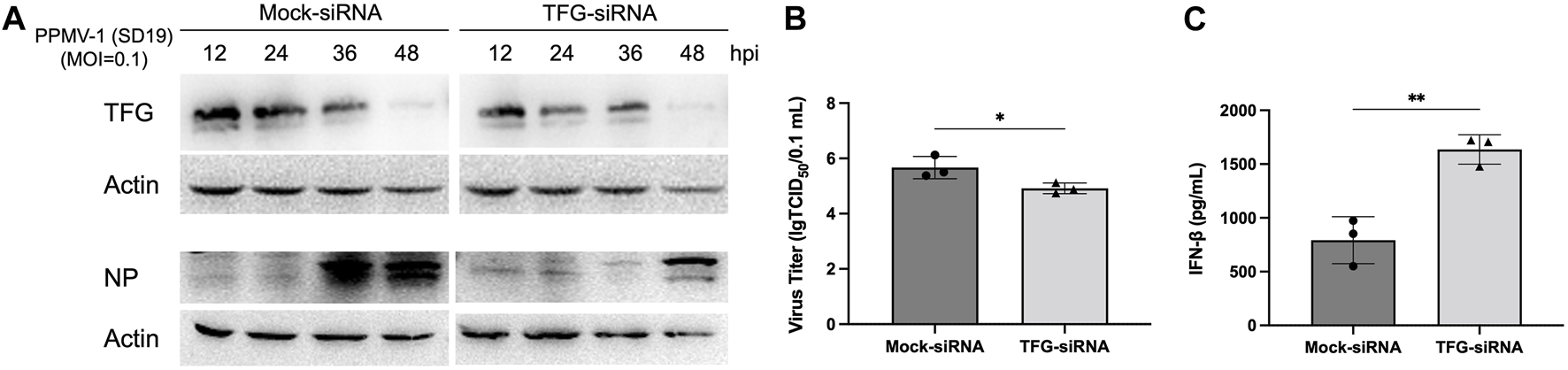
Knocking down TFG inhibited PPMV-1 replication by modulating the IFN-I pathway. (A) Control and TFG-knockdown Hela cells were infected with PPMV-1 (MOI = 0.1) for the indicated time. NP and TFG in cell lysate were detected by Western blotting. (B) In parallel, the culture supernatant of 36 h after PPMV-1 infection was subjected to TCID50 assay, to measure the released progeny virus. (C) Control and TFG-knockdown Hela cells were infected with PPMV-1 (MOI = 0.1) for 16 h, supernatants were subjected to an ELISA to detect IFN-β. The data represent the means ± SD of three independent experiments. Statistical analyses were performed using GraphPad Prism software (*, p < 0.05; **, p < 0.01).
Studies have shown that TFG can downregulate the RIG-I-mediated type I IFN response. Therefore, we also investigated whether TFG promotes viral replication via its inhibition of type I IFN signaling in the early stages of PPMV-1 infection. We depleted TFG using siRNAs in Hela cells and then infected the cells with PPMV-1 (MOI = 0.1). Compared with the controls, the expression of IFN-β in cellular supernatant in TFG knockdown cells was significantly increased (Figure 3C), indicating that TFG may affect virus replication by inhibiting the production of IFN-β.
TFG inhibits interferon response triggered by RIG-I and MDA5
To understand the exact role of TFG in the IFN-I signaling, we analyzed the effect of TFG on IFN-I activation. Hela cells were co-transfected with the TFG protein expression plasmid and the IFN-β activating reporter plasmid. The NS1 influenza virus protein expression plasmid, which is a potent IFN-I inhibitor, was used as the control. 2 μg/mL Poly (I:C) was transfected into the cells to induce IFN-β expression at the same time. The relative luciferase activity was measured 24 h later. As shown in Figure 4A, TFG inhibited activation of the Poly (I:C)-stimulated IFN response, as did the influenza virus NS1 protein, indicating that TFG protein is an IFN inhibitor. To detect the molecular mechanism by which TFG inhibits the IFN-I response, the TFG protein expression plasmid was co-transfected with protein molecules from different segments of the IFN-I pathway. The IFN-I reporter gene system was used to identify where TFG inhibits the IFN-I pathway. As shown in Figures 4B, C, TFG protein inhibited the IFN-I response stimulated by RIG-CARD and MDA5, indicating that IFN-I inhibition by TFG occurred in RIG-CARD and MDA5 or downstream of them.
FIGURE 4
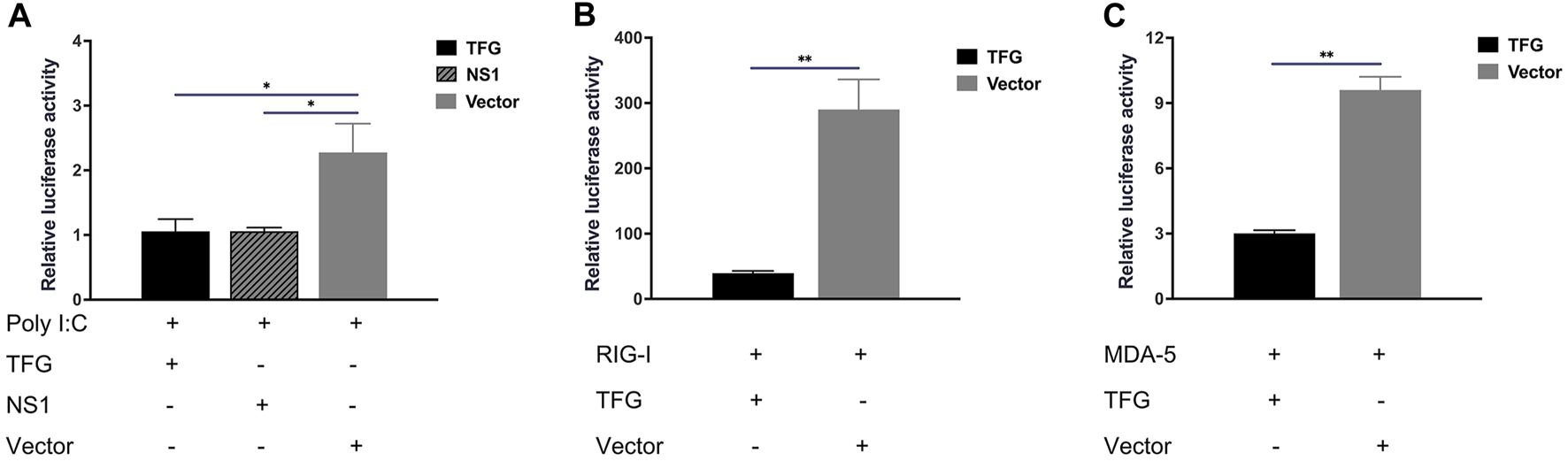
TFG inhibited the IFN-I response activated by RIG-I and MDA5. (A) Relative luciferase reporter gene expression levels under Poly (I:C) stimulation in HeLa cells transfected with TFG or NS1. Relative luciferase reporter gene expression levels in HeLa cells transfected with TFG and RIG-I (B) or MDA5 (C). The data represent the means ± SD of three independent experiments. Statistical analyses were performed using GraphPad Prism software (*, p < 0.05; **, p < 0.01).
TFG inhibits interferon activation by MAVS and TBK1 instead of IKKε and IRF3
To better understand how TFG interacts with the interferon system, HeLa cells were co-transfected with the MAVS and TFG plasmid, and the IFN-I reporter gene system was used for detection. That TFG inhibited the IFN-I response activated by MAVS protein indicates that IFN-I inhibition by TFG may occur by MAVS or a downstream protein in the pathway (Figure 5A). Next, TBK1 or IKKε was co-transfected with the TFG protein expression plasmid into HeLa cells. The results showed that TFG significantly inhibited the IFN-I response activated by TBK1 (Figure 5B), but did not inhibit the IFN-I response activated by IKKε (Figure 5C). IRF3 is the last step in the IFN-I pathway before activation. As shown in Figure 5D, after co-transfection of the TFG and IRF3 expression plasmid, the IFN-I reporting system showed that TFG did not inhibit the IRF3-activated IFN-I pathway. Instead, it indicated that TFG can only inhibit the activation of IFN-I in the pathway upstream of IRF3. These results show that TFG is likely to inhibit the IFN-I pathway by downregulating the TBK1 protein’s function.
FIGURE 5
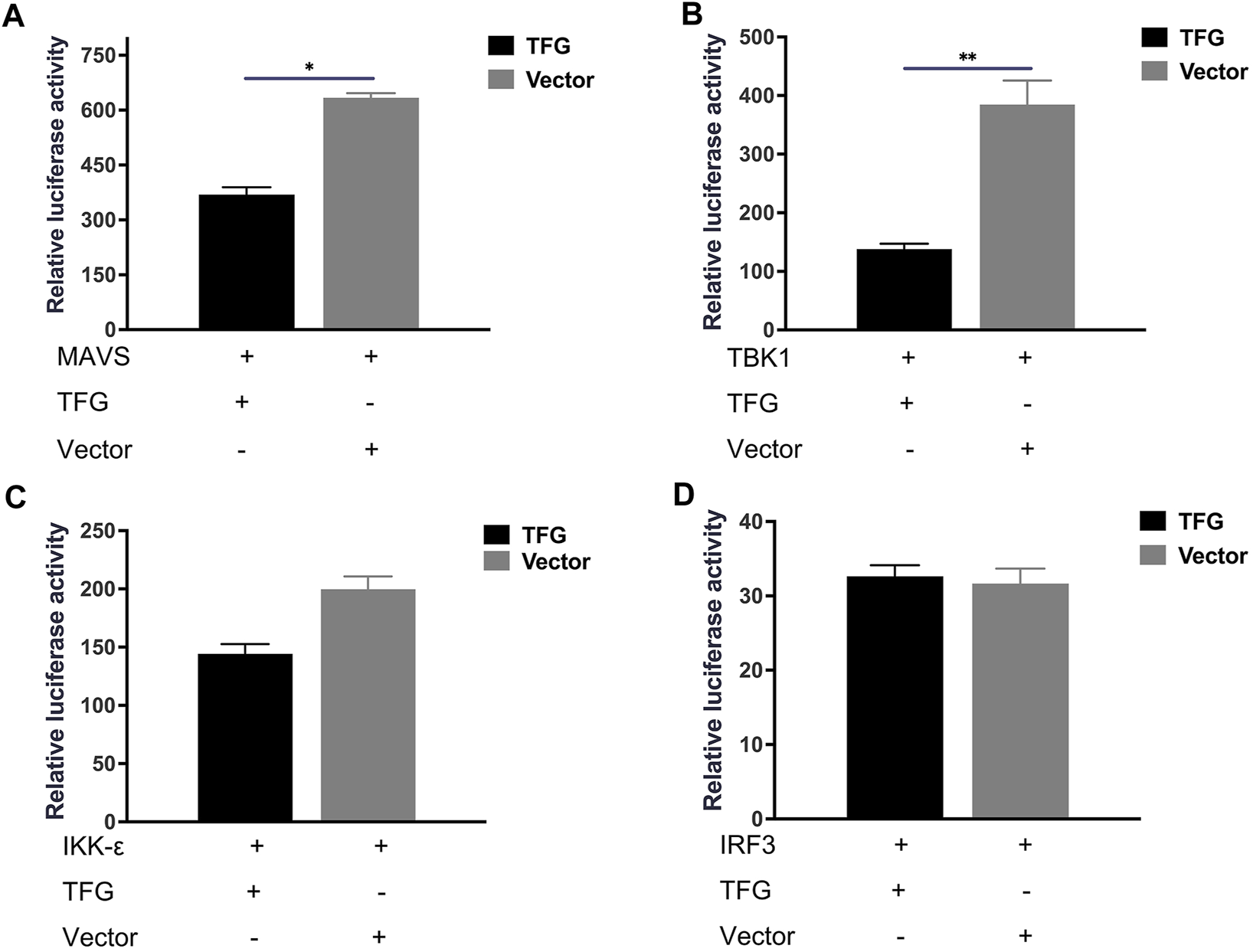
TFG inhibited interferon activation by MAVS and TBK1 instead of IKK and IRF3. Relative luciferase reporter gene expression levels in HeLa cells transfected with TFG and MAVS (A), TBK-1 (B), IKK-ε (C) or IRF3 (D). The data represent the means ± SD of three independent experiments. Statistical analyses were performed using GraphPad Prism software (*, p < 0.05; **, p < 0.01).
TFG inhibits phosphorylation of TBK1 and IRF3
To further explore how TFG inhibits TBK1, we first overexpressed TFG protein at different concentrations to investigate its effect on TBK1 protein expression levels. The results (Figure 6A) showed that TFG did not affect the total expression level of TBK1. Phosphorylation is a common post-translational modification of proteins and one of the most important modifications to regulate their functions. Phosphorylation of TBK1 protein is very important for its ability to activate interferon pathways. Therefore, we detected whether TFG protein could affect the phosphorylation of TBK1 protein. After overexpressing TFG protein, the phosphorylation level of the TBK1 protein was detected when the IFN pathway was activated. The results (Figure 6B) showed that the phosphorylation level of TBK1 was significantly reduced in the TFG overexpression group. This indicates that TFG protein may inhibit interferon activation by reducing the phosphorylation level of TBK1 protein. In the IFN pathway, TBK1 protein activates downstream IRF3 phosphorylation. After phosphorylation, IRF3 forms dimers and is transferred to the nucleus where it plays the role of a transcription factor to induce IFN-I transcription. To verify that the inhibition of TFG on TBK1 phosphorylation affected the phosphorylation of IRF3 protein, we overexpressed TFG to detect the level of phosphorylated IRF3 when the IFN pathway was activated. As shown in Figures 6C, D, after overexpressing TFG protein while the IFN pathway was activated, the phosphorylation level of IRF3 became downregulated, but the total protein level of IRF3 did not change, indicating that overexpression of TFG did not affect the protein level of IRF3, but inhibited the phosphorylation of IFR3. These results indicate that the TFG did not inhibit the IFN response activated by IRF3, but inhibited the phosphorylation of TBK1 protein, thereby inhibiting IFNs production.
FIGURE 6
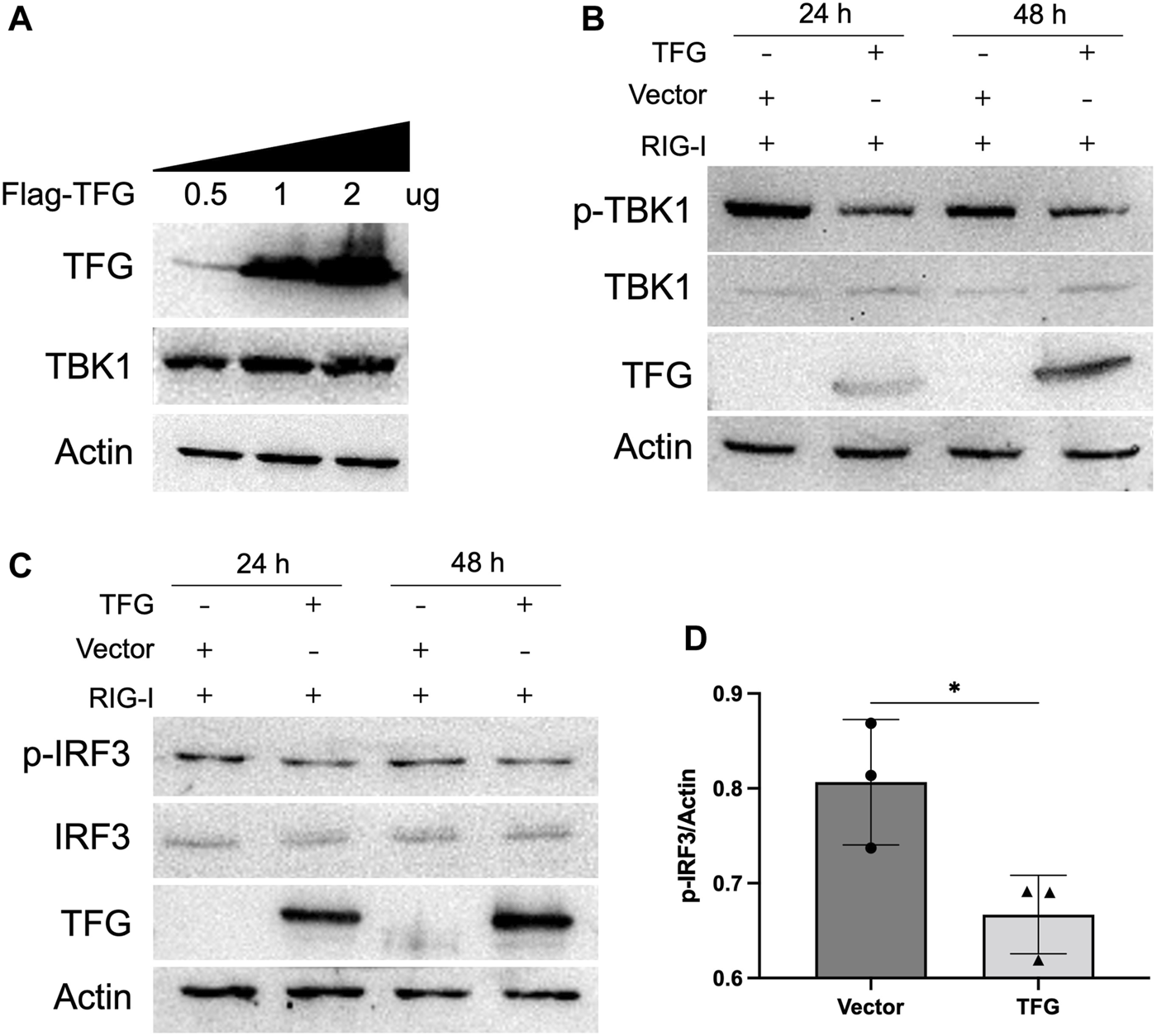
TFG inhibited the phosphorylation of TBK1 and IRF3. (A) The protein expression levels of TBK1 in HeLa cells with different concentrations of TFG were examsined by Western blotting. (B) The protein expression levels of p-TBK1 and TBK1 in HeLa cells at 24 and 48 h after TFG transfection were examined by Western blotting. (C,D) The protein expression levels of p-IRF3 and IRF3 in HeLa cells at 24 and 48 h after TFG transfection were examined by Western blotting. The relative levels of p-IRF3 to actin expression were determined by densitometry using Image J software version 1.8.0. The data represent the means ± SD of three independent experiments. Statistical analyses were performed using GraphPad Prism software (*, p < 0.05).
Discussion
The innate immune responses triggered by paramyxovirus infection have been extensively studied. However, a variety of strategies have evolved in paramyxoviruses to counter IFN synthesis in their hosts, such as controlling viral RNA synthesis, inhibiting cellular RNA sensors, blocking signaling kinase complexes, and suppressing IFN promoter activity (Parks and Alexander-Miller, 2013), all of which benefit virus replication. For instance, it is reported that APMV-1 hemagglutinin protein expression can upregulate the SAPK/JNK (stress-activated protein kinase/c-Jun N-terminal kinase) pathway, leading to transactivation of c-Jun and apoptosis signaling activation (Rajmani et al., 2015). The P gene of APMV-1 encodes three viral proteins via RNA editing, among which the V protein encodes an IFN antagonistic activity that inhibits cell apoptosis (Chu et al., 2018a; Chu et al., 2018b; Wang et al., 2018). While Sánchez-Aparicio et al. (2018) reported that the interaction between paramyxovirus V proteins and TRIM25 and RIG-I inhibits TRIM25-mediated ubiquitination of RIG-I, Qiu et al. (2016) reported that the C-terminal domain of the V protein encoded by APMV-1 can cause degradation of STAT1 and block IFN signal transduction. Sun et al. (2019) found that the V protein could target ubiquitination and degradation of MAVS through E3 ligase RNF5, resulting in the interruption of IFN signal transduction and the inhibition of IFN signaling activation. Lastly, Wang et al. (2019) found that the non-structural V protein in APMV-1 induced SOCS3 expression through the MEK/ERK signaling pathway, and facilitated viral replication.
As well as the paramyxovirus using its own protein to stop the host’s IFN pathway being activated, we found that the virus antagonized IFN by upregulating the inhibitor of the host’s IFN response during infection. The RIG-I-mediated antiviral IFN pathway is the host’s first barrier of defense against the virus. The RIG-I structure in host cells changes after sensing viral RNA, and the SPRY domain of TRIM25 interacts with the N-CARD region of RIG-I. This interaction leads to K63 ubiquitination of the CARD region of RIG-I, which activates RIG-I and then further recruits proteins such as MAVS to activate the downstream IFN pathway (Sanchez et al., 2016; Martín-Vicente et al., 2017; Rehwinkel and Gack, 2020). TFG can interact with TRIM25 upon viral infection and inhibit the reactions of IFNs (Lee et al., 2013a). Overexpression of TRIM 11 was found to enhance viral infectivity and inhibit RIG-I-mediated IFN-β production by targeting TBK1 signaling (Lee et al., 2013b). Here, we found that TFG protein was significantly upregulated during the early stage of PPMV-1 infection and IFN activation was inhibited, thus facilitating viral replication. However, in the late infection stages, TFG protein levels were significantly downregulated, possibly caused by autophagy and apoptosis of cells during this late stage (Liao et al., 2017; Cai et al., 2023). Lacking a self-contained metabolism network, viruses hijack the host’s metabolic resources for replication, which may inhibit the expression of some host proteins. We focused on the effect of TFG on IFN-I pathway in PPMV-1-infected cells. However, TFG is an evolutionarily conserved secretion system regulator protein, and its deletion may have an impact on the whole complex of events in infected cells, and affect viral infection through other ways, which is what we need to focus on in the future.
To further study the specific molecular mechanism whereby TFG inhibits IFN-β, we investigated the effect of TFG inhibition on IFN-β activation by different molecules in the IFN pathway. The results showed that TFG was able to inhibit IFN-β activation by RIG-CARD, MAVS and TBK1, but was unable to inhibit IFN-β activation by IRF3, indicating that TFG inhibits IFN activation by inhibiting the function of TBK1 protein. TBK1 activation can be regulated by ubiquitination, phosphorylation, kinase activity modulation and by preventing functional TBK1-containing complex formation (Zhao, 2013). Our further experiments showed that TFG did not affect total TBK1 protein expression levels but did inhibit the TBK1 phosphorylation level. Although studies have shown that TFG is required for the phosphorylation activation of TBK1(Khan et al., 2021), but our results are consistent with Lee’s research (Lee et al., 2013a). TBK1 is an important protein kinase that phosphorylates IRF3. After homo-dimerization, IRF3 translocates to the nucleus and drives IFN-β gene transcription (Bakshi et al., 2017). In addition to TBK1, IRF3 is also phosphorylated by IKKɛ. However, in the present study, the IFN-β promoter reported that TFG inhibited the TBK1-activated IFN reaction, instead of the IKKɛ activated IFN reaction, which further suggests that TFG has specificity for IFN-β activation by inhibiting TBK1 function. Husain et al. (2018) reported that TBK1 phosphorylation is important for TBK1 protein functioning. Inhibition of TBK1 phosphorylation by TFG protein, as was seen in our study, may affect the activation of a kinase complex comprising TBK1 and IKKε. This kinase complex phosphorylates the latent cytoplasmic transcription factor IRF3. However, more work is required to clarify how TFG regulates TBK1 phosphorylation during pathogenic viral infections.
In conclusion, we have shown that TFG is an important negative regulator, which targets TBK1 phosphorylation and inhibits IRF3 phosphorylation mediated by pTBK1. Consequently, TFG inhibits IFN-β production to the benefit of viral replication during the early stage of infection.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
YT retrieved literatures, performed research, and wrote the manuscript. JL proposed the topic and provided outline. RX provide methods and techniques. CY retrieved literatures and analyzed data. LL and SC revised the manuscript and reviewed the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2022E and CXGC2023F11).
Acknowledgments
We thank Sandra Cheesman, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Akira S. (2009). Pathogen recognition by innate immunity and its signaling. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci.85, 143–156. 10.2183/pjab.85.143
2
Bakshi S. Taylor J. Strickson S. McCartney T. Cohen P. (2017). Identification of TBK1 complexes required for the phosphorylation of IRF3 and the production of interferon β. Biochem. J.474, 1163–1174. 10.1042/BCJ20160992
3
Cai J. Wang S. Du H. Fan L. Yuan W. Xu Q. et al (2023). NDV-induced autophagy enhances inflammation through NLRP3/Caspase-1 inflammasomes and the p38/MAPK pathway. Vet. Res.54, 43. 10.1186/s13567-023-01174-w
4
Chu Z. Ma J. Wang C. Lu K. Li X. Liu H. et al (2018a). Newcastle disease virus V protein promotes viral replication in HeLa cells through the activation of MEK/ERK signaling. Viruses10, 489. 10.3390/v10090489
5
Chu Z. Wang C. Tang Q. Shi X. Gao X. Ma J. et al (2018b). Newcastle disease virus V protein inhibits cell apoptosis and promotes viral replication by targeting CacyBP/SIP. Front. Cell Infect. Microbiol.8, 304. 10.3389/fcimb.2018.00304
6
Faísca P. Desmecht D. (2007). Sendai virus, the mouse parainfluenza type 1: a longstanding pathogen that remains up-to-date. Res. Vet. Sci.82, 115–125. 10.1016/j.rvsc.2006.03.009
7
Ganar K. Das M. Sinha S. Kumar S. (2014). Newcastle disease virus: current status and our understanding. Virus Res.184, 71–81. 10.1016/j.virusres.2014.02.016
8
Ginting T. E. Suryatenggara J. Christian S. Mathew G. (2017). Proinflammatory response induced by newcastle disease virus in tumor and normal cells. Oncolytic Virother6, 21–30. 10.2147/OV.S123292
9
Gogoi P. Ganar K. Kumar S. (2017). Avian paramyxovirus: a brief review. Transbound. Emerg. Dis.64, 53–67. 10.1111/tbed.12355
10
Guo H. Liu X. Han Z. Shao Y. Chen J. Zhao S. et al (2013). Phylogenetic analysis and comparison of eight strains of pigeon paramyxovirus type 1 (PPMV-1) isolated in China between 2010 and 2012. Arch. Virol.158, 1121–1131. 10.1007/s00705-012-1572-8
11
Guo H. Liu X. Xu Y. Han Z. Shao Y. Kong X. et al (2014). A comparative study of pigeons and chickens experimentally infected with PPMV-1 to determine antigenic relationships between PPMV-1 and NDV strains. Vet. Microbiol.168, 88–97. 10.1016/j.vetmic.2013.11.002
12
Hanna M. G. Block S. Frankel E. B. Hou F. Johnson A. Yuan L. et al (2017). TFG facilitates outer coat disassembly on COPII transport carriers to promote tethering and fusion with ER-Golgi intermediate compartments. Proc. Natl. Acad. Sci. U. S. A.114, E7707–E7716. 10.1073/pnas.1709120114
13
He Y. Lu B. Dimitrov K. M. Liang J. Chen Z. Zhao W. et al (2020). Complete genome sequencing, molecular epidemiological, and pathogenicity analysis of pigeon paramyxoviruses type 1 isolated in guangxi, China during 2012-2018. Viruses12, 366. 10.3390/v12040366
14
Hou F. Sun L. Zheng H. Skaug B. Jiang Q.-X. Chen Z. J. (2011). MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell146, 448–461. 10.1016/j.cell.2011.06.041
15
Hu Z. Hu J. Hu S. Liu X. Wang X. Zhu J. et al (2012). Strong innate immune response and cell death in chicken splenocytes infected with genotype VIId Newcastle disease virus. Virol. J.9, 208. 10.1186/1743-422X-9-208
16
Husain S. Kumar V. Hassan M. I. (2018). Phosphorylation-induced changes in the energetic frustration in human Tank binding kinase 1. J. Theor. Biol.449, 14–22. 10.1016/j.jtbi.2018.04.016
17
Johnson A. Bhattacharya N. Hanna M. Pennington J. G. Schuh A. L. Wang L. et al (2015). TFG clusters COPII-coated transport carriers and promotes early secretory pathway organization. EMBO J.34, 811–827. 10.15252/embj.201489032
18
Kaleta E. F. Alexander D. J. Russell P. H. (1985). The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons. Avian Pathol.14, 553–557. 10.1080/03079458508436258
19
Khan K. A. Marineau A. Doyon P. Acevedo M. Durette É. Gingras A.-C. et al (2021). TRK-fused gene (TFG), a protein involved in protein secretion pathways, is an essential component of the antiviral innate immune response. PLoS Pathog.17, e1009111. 10.1371/journal.ppat.1009111
20
Kommers G. D. King D. J. Seal B. S. Brown C. C. (2001). Virulence of pigeon-origin newcastle disease virus isolates for domestic chickens. Avian Dis.45, 906–921. 10.2307/1592870
21
Lee N.-R. Shin H.-B. Kim H.-I. Choi M.-S. Inn K.-S. (2013a). Negative regulation of RIG-I-mediated antiviral signaling by TRK-fused gene (TFG) protein. Biochem. Biophys. Res. Commun.437, 168–172. 10.1016/j.bbrc.2013.06.061
22
Lee S.-H. Kim J. S. Jun H.-K. Lee H.-R. Lee D. Choi B.-K. (2009). The major outer membrane protein of a periodontopathogen induces IFN-beta and IFN-stimulated genes in monocytes via lipid raft and TANK-binding kinase 1/IFN regulatory factor-3. J. Immunol.182, 5823–5835. 10.4049/jimmunol.0802765
23
Lee Y. Song B. Park C. Kwon K.-S. (2013b). TRIM11 negatively regulates IFNβ production and antiviral activity by targeting TBK1. PLoS One8, e63255. 10.1371/journal.pone.0063255
24
Liao Y. Wang H.-X. Mao X. Fang H. Wang H. Li Y. et al (2017). RIP1 is a central signaling protein in regulation of TNF-α/TRAIL mediated apoptosis and necroptosis during newcastle disease virus infection. Oncotarget8, 43201–43217. 10.18632/oncotarget.17970
25
Liu H. Wang Z. Son C. Wang Y. Yu B. Zheng D. et al (2006). Characterization of pigeon-origin newcastle disease virus isolated in China. Avian Dis.50, 636–640. 10.1637/7618-042606R1.1
26
Liu M. Qu Y. Wang F. Liu S. Sun H. (2015). Genotypic and pathotypic characterization of newcastle disease virus isolated from racing pigeons in China. Poult. Sci.94, 1476–1482. 10.3382/ps/pev106
27
Martín-Vicente M. Medrano L. M. Resino S. García-Sastre A. Martínez I. (2017). TRIM25 in the regulation of the antiviral innate immunity. Front. Immunol.8, 1187. 10.3389/fimmu.2017.01187
28
Meng S. Zhou Z. Chen F. Kong X. Liu H. Jiang K. et al (2012). Newcastle disease virus induces apoptosis in cisplatin-resistant human lung adenocarcinoma A549 cells in vitro and in vivo. Cancer Lett.317, 56–64. 10.1016/j.canlet.2011.11.008
29
Miranda C. Roccato E. Raho G. Pagliardini S. Pierotti M. A. Greco A. (2006). The TFG protein, involved in oncogenic rearrangements, interacts with TANK and NEMO, two proteins involved in the NF-κB pathway. J. Cell Physiol.208, 154–160. 10.1002/jcp.20644
30
Parks G. D. Alexander-Miller M. A. (2013). Paramyxovirus activation and inhibition of innate immune responses. J. Mol. Biol.425, 4872–4892. 10.1016/j.jmb.2013.09.015
31
Peotter J. Kasberg W. Pustova I. Audhya A. (2019). COPII-mediated trafficking at the ER/ERGIC interface. Traffic20, 491–503. 10.1111/tra.12654
32
Qiu X. Fu Q. Meng C. Yu S. Zhan Y. Dong L. et al (2016). Newcastle disease virus V protein targets phosphorylated STAT1 to block IFN-I signaling. PLoS One11, e0148560. 10.1371/journal.pone.0148560
33
Rajmani R. S. Gandham R. K. Gupta S. K. Sahoo A. P. Singh P. K. Kumar R. et al (2015). HN protein of newcastle disease virus induces apoptosis through SAPK/JNK pathway. Appl. Biochem. Biotechnol.177, 940–956. 10.1007/s12010-015-1788-7
34
Rehwinkel J. Gack M. U. (2020). RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol.20, 537–551. 10.1038/s41577-020-0288-3
35
Rima B. Balkema-Buschmann A. Dundon W. G. Duprex P. Easton A. Fouchier R. et al (2019). ICTV virus taxonomy profile: paramyxoviridae. J. Gen. Virol.100, 1593–1594. 10.1099/jgv.0.001328
36
Sánchez-Aparicio M. T. Feinman L. J. García-Sastre A. Shaw M. L. (2018). Paramyxovirus V proteins interact with the RIG-I/TRIM25 regulatory complex and inhibit RIG-I signaling. J. Virol.92, 01960–e2017. 10.1128/JVI.01960-17
37
Sanchez J. G. Chiang J. J. Sparrer K. M. J. Alam S. L. Chi M. Roganowicz M. D. et al (2016). Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep.16, 1315–1325. 10.1016/j.celrep.2016.06.070
38
Sawatsky B. Cattaneo R. von Messling V. (2018). Canine distemper virus spread and transmission to naive ferrets: selective pressure on signaling lymphocyte activation molecule-dependent entry. J. Virol.92, 00669–e718. 10.1128/JVI.00669-18
39
Seth R. B. Sun L. Ea C.-K. Chen Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell122, 669–682. 10.1016/j.cell.2005.08.012
40
Śmietanka K. Olszewska M. Domańska-Blicharz K. Bocian A. L. Minta Z. (2014). Experimental infection of different species of birds with pigeon paramyxovirus type 1 virus--evaluation of clinical outcomes, viral shedding, and distribution in tissues. Avian Dis.58, 523–530. 10.1637/10769-011514-Reg.1
41
Sun Y. Zheng H. Yu S. Ding Y. Wu W. Mao X. et al (2019). Newcastle disease virus V protein degrades mitochondrial antiviral signaling protein to inhibit host type I interferon production via E3 ubiquitin ligase RNF5. J. Virol.93, 00322–e419. 10.1128/JVI.00322-19
42
Thibault P. A. Watkinson R. E. Moreira-Soto A. Drexler J. F. Lee B. (2017). Zoonotic potential of emerging paramyxoviruses: knowns and unknowns. Adv. Virus Res.98, 1–55. 10.1016/bs.aivir.2016.12.001
43
Tian Y. Xue R. Yang W. Li Y. Xue J. Zhang G. (2020). Characterization of ten paramyxovirus type 1 viruses isolated from pigeons in China during 1996-2019. Vet. Microbiol.244, 108661. 10.1016/j.vetmic.2020.108661
44
Wang C. Chu Z. Liu W. Pang Y. Gao X. Tang Q. et al (2018). Newcastle disease virus V protein inhibits apoptosis in DF-1 cells by downregulating TXNL1. Vet. Res.49, 102. 10.1186/s13567-018-0599-6
45
Wang X. Jia Y. Ren J. Huo N. Liu H. Xiao S. et al (2019). Newcastle disease virus nonstructural V protein upregulates SOCS3 expression to facilitate viral replication depending on the MEK/ERK pathway. Front. Cell Infect. Microbiol.9, 317. 10.3389/fcimb.2019.00317
46
Wang X. Ren S. Wang X. Wang C. Y. Fan M. Jia Y. et al (2017). Genomic characterization of a wild-bird-origin pigeon paramyxovirus type 1 (PPMV-1) first isolated in the northwest region of China. Arch. Virol.162, 749–761. 10.1007/s00705-016-3156-5
47
Wynne C. Lazzari E. Smith S. McCarthy E. M. Ní Gabhann J. Kallal L. E. et al (2014). TRIM68 negatively regulates IFN-β production by degrading TRK fused gene, a novel driver of IFN-β downstream of anti-viral detection systems. PLoS One9, e101503. 10.1371/journal.pone.0101503
48
Xie P. Chen L. Zhang Y. Lin Q. Ding C. Liao M. et al (2020). Evolutionary dynamics and age-dependent pathogenesis of sub-genotype VI.2.1.1.2.2 PPMV-1 in pigeons. Viruses12, E433. 10.3390/v12040433
49
Yamamotoya T. Nakatsu Y. Kushiyama A. Matsunaga Y. Ueda K. Inoue Y. et al (2017). Trk-fused gene (TFG) regulates pancreatic β cell mass and insulin secretory activity. Sci. Rep.7, 13026. 10.1038/s41598-017-13432-x
50
Yoneyama M. Kikuchi M. Natsukawa T. Shinobu N. Imaizumi T. Miyagishi M. et al (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol.5, 730–737. 10.1038/ni1087
51
Yu X. Luo Y. Wang J. Shu B. Jiang W. Liu S. et al (2022). A molecular, epidemiological and pathogenicity analysis of pigeon paramyxovirus type 1 viruses isolated from live bird markets in China in 2014-2021. Virus Res.318, 198846. 10.1016/j.virusres.2022.198846
52
Zeltina A. Bowden T. A. Lee B. (2016). Emerging paramyxoviruses: receptor tropism and zoonotic potential. PLoS Pathog.12, e1005390. 10.1371/journal.ppat.1005390
53
Zhan T. Lu X. He D. Gao X. Chen Y. Hu Z. et al (2022). Phylogenetic analysis and pathogenicity assessment of pigeon paramyxovirus type 1 circulating in China during 2007-2019. Transbound. Emerg. Dis.69, 2076–2088. 10.1111/tbed.14215
54
Zhang D. Ding Z. Xu X. (2023). Pathologic mechanisms of the newcastle disease virus. Viruses15, 864. 10.3390/v15040864
55
Zhao J. Liu C. Zhang J. Huang X. Zhang G. (2019). Cytokine expression in chicken embryo fibroblasts in response to infection with virulent or lentogenic avian avulavirus 1 (AAvV-1). Microb. Pathog.133, 103556. 10.1016/j.micpath.2019.103556
56
Zhao W. (2013). Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett.587, 542–548. 10.1016/j.febslet.2013.01.052
Summary
Keywords
TFG, IFN response, phosphorylation, TBK1, PPMV-1
Citation
Tian Y, Xue R, Yu C, Liu L, Chen S and Lv J (2023) The TRK-fused gene negatively regulates interferon signaling by inhibiting TBK1 phosphorylation during PPMV-1 infection. Acta Virol. 67:11607. doi: 10.3389/av.2023.11607
Received
24 May 2023
Accepted
19 October 2023
Published
03 November 2023
Volume
67 - 2023
Edited by
Ivana Nemčovičová, Slovak Academy of Sciences, Slovakia
Reviewed by
Jitka Forstová, Charles University, Czechia
Peter Kabát, Institute of Virology (SAS), Slovakia
Katarina Lopusna, Slovak Academy of Sciences, Slovakia
Updates
Copyright
© 2023 Tian, Xue, Yu, Liu, Chen and Lv.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junfeng Lv, nkyljf@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.