Abstract
The genus Potyvirus (the family Potyviridae) is the largest group of plant-infecting viruses transmitted by aphids. Through high-throughput RNA sequencing analysis of asymptomatic samples of Aconitum carmichaelii, a significant medicinal herb in Asia, we identified the genome sequences of two RNA viruses, tentatively named Aconitum potyvirus 1 (AcoPV1) and Aconitum potyvirus 2 (AcoPV2). The genomes of AcoPV1 and AcoPV2 encode polyproteins composed of 3,069 and 3,054 amino acids, respectively. Sequence comparisons and phylogenetic analyses established that AcoPV1 and AcoPV2 represent unique, novel members within the genus Potyvirus. The estimated RNA polymerase slippage rates at the GAAAAAA motif, responsible for the production of P3N-PIPO or P3N-ALT trans-frame fusion proteins, were 0.79% in AcoPV1 and 1.38% in AcoPV2. The RNA reads of AcoPV1 and AcoPV2 were predominantly found in the leaf and flower tissues, indicating potential feeding preferences of vectors for these viruses. These findings demonstrate the effectiveness of high-throughput RNA sequencing in not only uncovering novel potyviruses, but also in elucidating their genomic dynamics within host plants.
Introduction
The genus Potyvirus, belonging to the family Potyviridae, represents the largest genus of plant viruses, containing 201 species approved by the International Committee on Taxonomy of Viruses1 (last accessed 27 June, 2023) (Inoue-Nagata et al., 2022). Potyviruses have positive-sense single-stranded RNA genomes and encode an open reading frame (ORF) for a large polyprotein. The potyvirus polyprotein is proteolytically processed to yield ten mature peptides, named P1 protease (P1), helper component-protease (HC-Pro), P3 protein (P3), 6-kilodalton (kDa) peptide 1 (6K1), cylindrical inclusion protein (CI), 6-kDa peptide 2 (6K2), viral protein genome-linked (VPg), nuclear inclusion-a protease (NIa-Pro), nuclear inclusion-b protein (NIb), and capsid protein (CP), in the sequence from N- to C-terminus (Goh and Hahn, 2021; Pasin et al., 2022). The viral protease P1, HC-Pro, and NIa-Pro recognize nine cleavage sites within the polyprotein: P1 and HC-Pro cleaves the P1/HC-Pro and HC-Pro/P3 junctions respectively, while NIa-Pro processes the other seven junctions (P3/6K1, 6K1/CI, CI/6K2, 6K2/VPg, VPg/NIa-Pro, NIa-Pro/NIb, and NIb/CP) (Adams et al., 2005a; Goh and Hahn, 2021). The NIb protein is an RNA-dependent RNA polymerase (RdRp) and plays a vital role in viral replication.
Potyviruses and other members of the family Potyviridae carry an additional small ORF, which is referred to as the pretty interesting Potyviridae ORF (PIPO) (Chung et al., 2008; Park et al., 2017; Choi et al., 2021). During the replication of the viral genome, the viral RdRp occasionally slips at a conserved sequence of GAAAAAA (GA6) or a similar motif within the P3 coding region, leading to the insertion of an extra adenine (A) residue into the GA6 sequence (Olspert et al., 2015; Rodamilans et al., 2015; Choi et al., 2021). The efficiency of the A residue insertion has been estimated to be between 0.8% and 2.1% in several potyviruses (Olspert et al., 2015; Rodamilans et al., 2015; Olspert et al., 2016; Untiveros et al., 2016). When these RNA polymerase slippage products are translated, a trans-frame fusion protein known as P3N-PIPO, which comprises the N-terminal region of P3 and a PIPO-derived peptide, is produced (Chung et al., 2008; Olspert et al., 2015). The P3N-PIPO protein plays a crucial role in facilitating the movement of potyvirus genomic RNAs from cell to cell (Wen and Hajimorad, 2010; Wang, 2021). An A residue can also be deleted from the GA6 sequence, leading to the production of an alternative trans-frame fusion protein called P3N-ALT, which also plays a role in the movement of potyvirus genomic RNAs between cells (Hagiwara-Komoda et al., 2016).
Native plants are often associated with viruses that result in latent infections without exhibiting noticeable disease symptoms (Ilyas et al., 2022; Rosario et al., 2022; Cruz et al., 2023). The advent of high-throughput RNA sequencing (RNA-seq) has provided a tool for characterizing these viruses (Nibert et al., 2016; Bejerman and Debat, 2022). Numerous novel RNA virus genome sequences have been identified within assembled transcriptome contigs from diverse plant RNA-seq datasets (Goh et al., 2021; Park et al., 2021; Shin et al., 2022a; Shin et al., 2022b; Choi et al., 2022; Choi et al., 2023). In this study, we report discovery of two novel potyvirus genome sequences by high-throughput RNA-seq of Aconitum carmichaelii tissue samples.
Materials and methods
High-throughput RNA-seq data
In this study, we analyzed RNA-seq data originally collected from four tissue samples (flower, bud, leaf, and root) of a three-year-old A. carmichaelii plant with the aim of identifying key genes involved in the biosynthesis of diterpene alkaloids (Rai et al., 2017). The plant, which was cultivated under natural conditions at a research center in Tsukuba, Japan, did not show any symptoms of viral disease. The RNA-seq data are available in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under the accession numbers SRR6225420–SRR6225423. High-quality A. carmichaelii RNA-seq data were obtained by filtering raw data using sickle (version 1.332) with the parameter “-q 30 -l 55.” Transcriptome contigs were generated by assembling high-quality reads using the SPAdes Genome Assembler (version 3.15.43) with the parameter “--rnaviral” (Bushmanova et al., 2019).
Viral genome identification and annotation
Putative viral genome contigs were initially identified by comparing A. carmichaelii transcriptome contigs with known viral RdRp domain sequences extracted from the Pfam database (release 35.04) using DIAMOND (version v2.0.14.1525) (Buchfink et al., 2021). Accession numbers of the Pfam RdRp families used for preparation of known viral RdRp domains are PF00602, PF00603, PF00604, PF00680, PF00946, PF00972, PF00978, PF00998, PF02123, PF03431, PF04196, PF04197, PF05273, PF05788, PF05919, PF06317, PF06478, PF07925, PF08467, and PF12426. A polyprotein ORF of a putative potyvirus genome contig was predicted using the NCBI ORFfinder6. Protease cleavage sites of a polyprotein were identified by a sequence comparison with polyproteins of closely related viruses. Pairwise sequence identities were calculated using FASTA (version 36.3.8h7). Sequence logo representations were prepared using WebLogo (version 38) (Schneider and Stephens, 1990; Crooks et al., 2004).
RNA polymerase slippage analysis
High-quality reads were mapped to potyviral genome contig sequences using BWA (version 0.7.17-r1194-dirty9) with the BWA-MEM algorithm (Li and Durbin, 2009). RNA-seq reads that span a putative RNA polymerase slippage site in the P3 coding region were extracted, and the frequency of insertions or deletions of A residues was determined.
Phylogenetic analysis
MAFFT (version 7.47510), with the parameter “--auto,” was utilized for creating a multiple alignment of potyviral polyprotein sequences (Nakamura et al., 2018). Information-rich segments from the multiple sequence alignment were extracted using trimAl (version 1.4.rev2211) with the parameter “-automated1” (Capella-Gutierrez et al., 2009). The maximum likelihood phylogenetic tree was constructed using IQ-TREE (version 2.2.012) with the “-B 1000” parameter (Minh et al., 2020). The resulting phylogenetic tree was visualized using MEGA (version 11.0.1313) (Kumar et al., 2018).
Results and discussion
Identification of novel potyvirus genomes
RNA-seq reads previously generated from four tissue types (flower, bud, leaf, and root) of a three-year-old A. carmichaelii plant were assembled into contigs (Rai et al., 2017). Two of these contigs, with lengths of 9,492 and 9,441 bp respectively, exhibited significant sequence similarities to RdRp sequences of known potyviruses. They contained a full-length coding region for potyviral polyproteins, although the completeness of the genomes should be verified through a molecular method such as rapid amplification of cDNA ends. The predicted polyprotein ORFs had lengths of 9,210 and 9,165 bp, encoding 3,069 and 3,054 aa polyproteins, respectively.
BLAST searches of the NCBI protein database revealed that the two putative potyvirus genome contigs identified in the A. carmichaelii transcriptome were most similar to tulip breaking virus (TBV), a member of the genus Potyvirus, with approximately 56% amino acid sequence identity of polyprotein sequences (Wylie et al., 2019). The species demarcation criteria for potyviruses are generally accepted as <76% nucleotide identity of the large ORF and <82% amino acid identity of the polyprotein (Adams et al., 2005b; Inoue-Nagata et al., 2022). Therefore, the two newly identified viruses can be considered novel and distinct from previously known potyviruses. The second most closely related known potyvirus was lily mottle virus (LMoV), with around 53% amino acid sequence identity of polyprotein sequences (Zheng et al., 2003).
The polyprotein ORFs of the two newly identified potyvirus genome contigs exhibited a nucleotide sequence identity of 66%, while the amino acid sequence identity of polyprotein sequences was 69%. According to the aforementioned demarcation criteria, the two genome sequences were considered to originate from distinct potyviruses and were tentatively named Aconitum potyvirus 1 (AcoPV1, 9,492 bp) and Aconitum potyvirus 2 (AcoPV2, 9,441 bp). The AcoPV1 and AcoPV2 sequences were deposited in the NCBI GenBank under accession numbers OP271473 and OP271474, respectively.
The large polyprotein of a potyvirus is processed into ten mature peptides (P1, HC-Pro, P3, 6K1, CI, 6K2, VPg, NIa-Pro, NIb, and CP) by the three potyvirus proteases (P1, HC-Pro, and NIa-Pro) (Adams et al., 2005a; Goh and Hahn, 2021; Pasin et al., 2022). Cleavage sites and boundaries of mature peptides were predicted in the AcoPV1 and AcoPV2 polyproteins based on sequence similarities with mature proteins of closely related potyviruses, including TBV and LMoV (Figure 1). The polyprotein sequences of AcoPV1 and AcoPV2 surrounding the cleavage sites matched previously deduced consensus sequences for the potyvirus protease recognition sites (Adams et al., 2005a; Goh and Hahn, 2021).
FIGURE 1
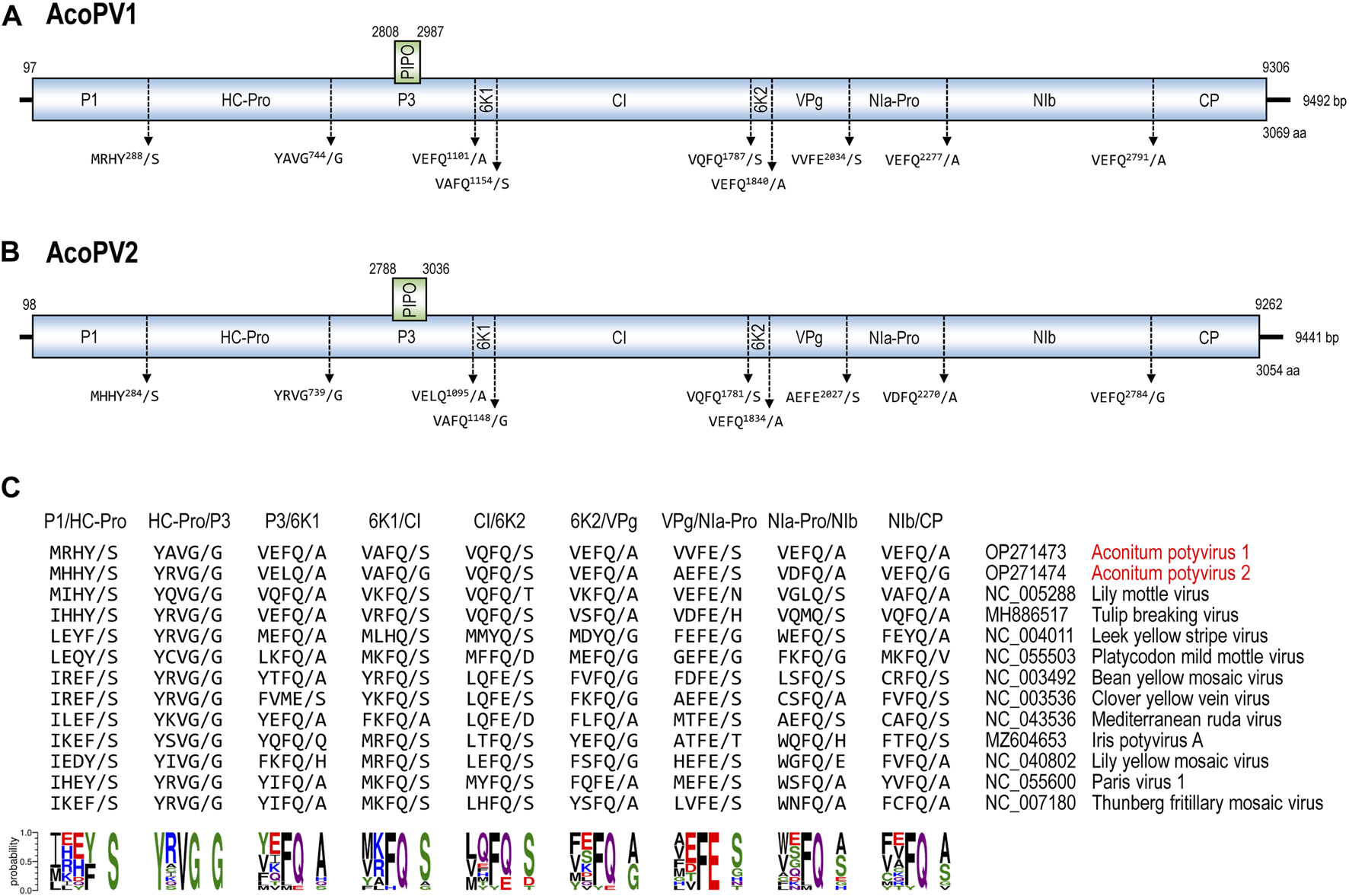
The genomic structure of AcoPV1 and AcoPV2. (A, B) The diagrams illustrate the genomic contigs of AcoPV1 (A) and AcoPV2 (B). The ORFs for the large polyprotein and PIPO are depicted as boxes, with their coordinates indicated above. Predicted cleavage sites are displayed below the polyprotein, with the slash (“/”) marking the cleaved bond. (C) The five amino acid residues at the nine cleavage sites of the AcoPV1, AcoPV2, and 11 closely related potyvirus polyprotein sequences are presented. At the bottom, sequence logo representations derived from the frequency of each amino acid residue at each position are shown.
All members of the family Potyviridae are known to have a secondary ORF, termed PIPO, which is situated in the P3 coding region, and is in frame −1 relative to the P3 (Yang et al., 2021; Inoue-Nagata et al., 2022). The PIPO ORF is expressed into a trans-frame fusion protein, known as P3N-PIPO, due to the insertion of an A residue at the slippery GA6 motif during replication (Chung et al., 2008). In the genomes of AcoPV1 and AcoPV2, the GA6 motifs were located at positions 2,802–2,808 and 2,782–2,788, respectively. The PIPO ORFs of AcoPV1 and AcoPV2 encode 59 and 82 amino acids, respectively. No additional GA6 motif other than that in the P3 coding region was found in AcoPV1 and AcoPV2 sequences.
RNA polymerase slippages of AcoPV1 and AcoPV2
The GA6 motif within the P3 coding region of potyviruses induces rare RNA polymerase slippages during replication, resulting in the crucial insertion of an A residue for P3N-PIPO protein production (Olspert et al., 2015; Rodamilans et al., 2015; Hagiwara-Komoda et al., 2016; Olspert et al., 2016; Untiveros et al., 2016). The RNA polymerase slippage rate can be estimated by counting RNA-seq reads originating from the slippage products in the GA6 motif. To detect and quantify the slippage products of the AcoPV1 and AcoPV2, RNA-seq reads spanning the GA6 motifs were extracted and examined (Figure 2).
FIGURE 2
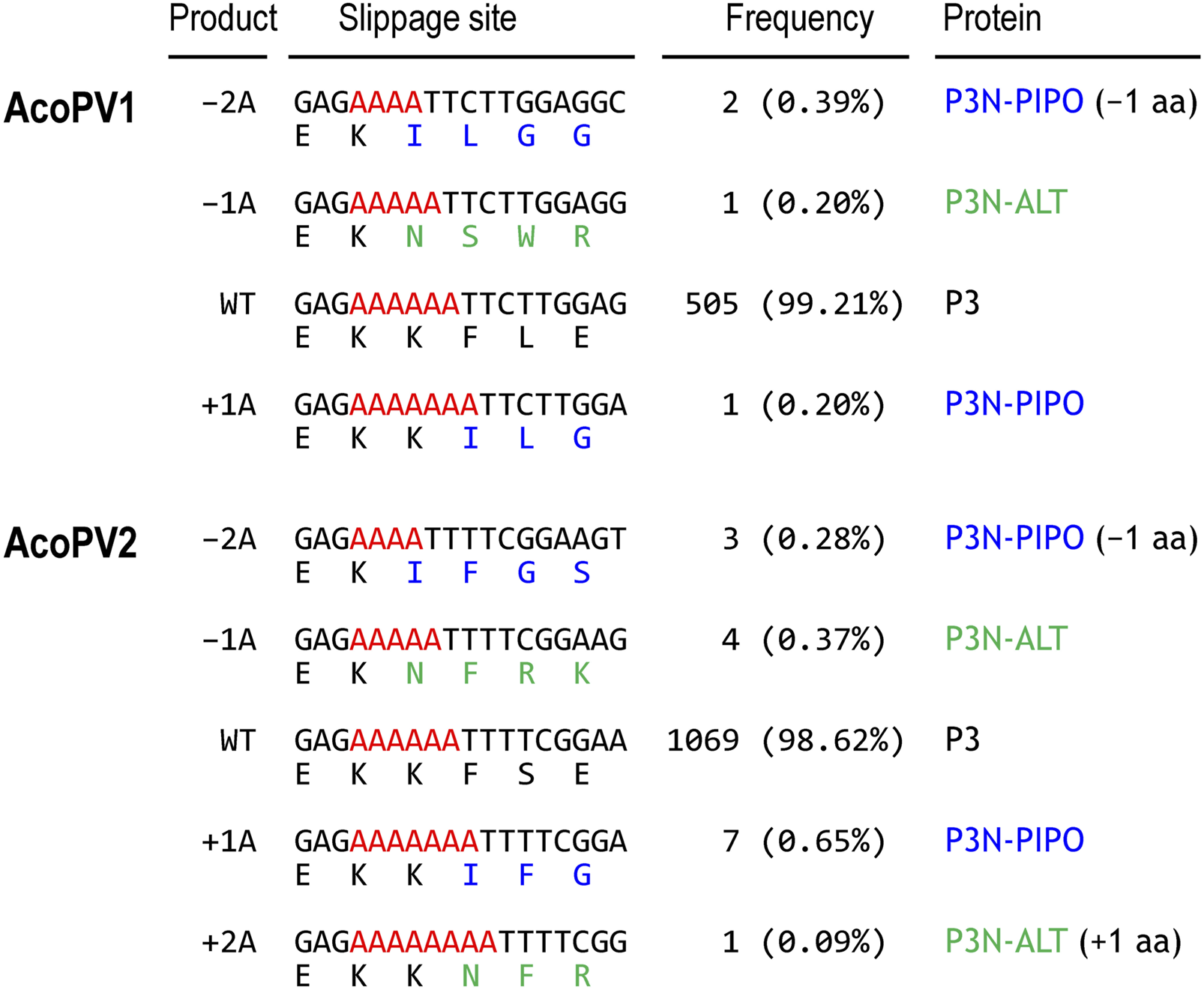
RNA polymerase slippages at the GA6 motif. Frequencies of slippage products generated by potential RNA polymerase slippage events in the AcoPV1 (top) and AcoPV2 (bottom) are presented. These slippage products include double-A deletion (–2A), single-A deletion (–1A), single-A insertion (+1A), and double-A insertion (+2A), compared to the wild type (WT). Adenine residues of the GA6 motif are highlighted in red. Amino acid sequences are color-coded to indicate residues from frame 0 (black, P3), frame −1 (blue, P3N-PIPO), and frame +1 (green, P3N-ALT).
A total of 509 and 1,084 RNA-seq reads were mapped to the GA6 motif in AcoPV1 and AcoPV2, respectively. Among these, 505 (99.21%) for AcoPV1 and 1,069 (98.62%) for AcoPV2 matched the original unmodified (wild type) sequence. There was one read for AcoPV1 (0.20%) and seven reads for AcoPV2 (0.65%) that potentially originated from the single-A insertion or +1A product, which is essential for P3N-PIPO protein production.
Interestingly, we also observed RNA-seq reads corresponding to other insertion or deletion products. One of these slippage products, the single-A deletion or –1A product, might lead to the production of another trans-frame fusion protein, known as P3N-ALT, as previously reported in clover yellow vein virus (ClYVV) (Hagiwara-Komoda et al., 2016). In AcoPV1, one read (0.20%) was identified as the –1A product, potentially encoding P3N-ALT, if present. The –1A product in AcoPV1 results in the addition of 12 amino acids after the GA6 motif. In the case of AcoPV2, four reads (0.37%) were identified as the –1A product, which could add five amino acids after the GA6 motif. Additionally, we also observed other slippage products, including double-A deletion (–2A) or double-A insertion (+2A) products. These slippage products may be translated to produce P3N-ALT with a single amino acid deletion or P3N-PIPO with a single amino acid insertion, respectively.
As a result, we estimated the rate of RNA polymerase slippage, potentially leading to the production of P3N-PIPO or P3N-ALT, to be 0.79% in AcoPV1 and 1.38% in AcoPV2. It is worth noting that the number of RNA-seq reads originating from slippage products is too low to accurately measure their RNA polymerase slippage rates.
Phylogenetic positions of AcoPV1 and AcoPV2
To determine the phylogenetic positions of AcoPV1 and AcoPV2 among known potyviruses, we retrieved polyprotein sequences of known potyviruses from the NCBI protein database. The pairwise identities of polyprotein sequences of AcoPV1, AcoPV2, and other potyviruses are presented in Supplementary Table S1. A maximum likelihood phylogenetic tree was inferred from a multiple sequence alignment that included AcoPV1, AcoPV2, and known polyprotein sequences (Figure 3).
FIGURE 3
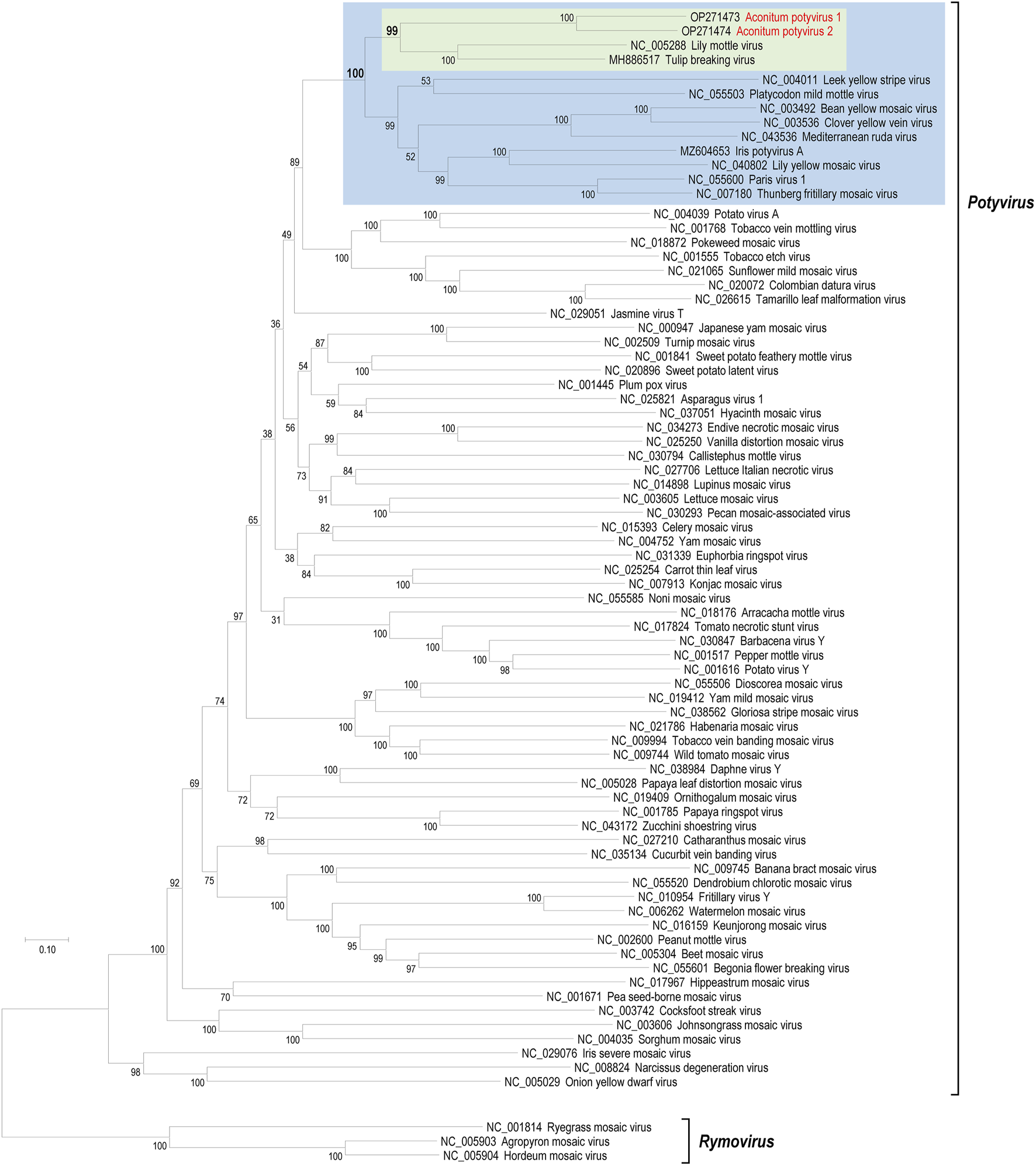
Phylogenetic positions of AcoPV1 and AcoPV2. The maximum likelihood phylogenetic tree was inferred from multiple alignment of selected potyvirus polyprotein sequences. The potyviruses most similar to AcoPV1 and AcoPV2 were lily mottle virus (LMoV) and tulip breaking virus (TBV), which are highlighted in the green box. AcoPV1, AcoPV2, LMoV, TBV, and other nine potyviruses formed a strong subclade, marked with a blue box, within the genus Potyvirus. The genus Rymovirus, represented by three rymoviruses, was used as an outgroup to root the genus Potyvirus.
Both AcoPV1 and AcoPV2 cluster together into a subclade, supported by a 100% bootstrap value, distinct from the known potyviruses. The clade that includes TBV and LMoV is the most closely related known potyvirus group to AcoPV1 and AcoPV2. Together, the AcoPV1/AcoPV2 and TBV/LMoV subclades form a single clade, supported by a 99% bootstrap value (green box in Figure 3).
The next most closely related clade comprises nine potyviruses: leek yellow stripe virus, Platycodon mild mottle virus, bean yellow mosaic virus, ClYVV, Mediterranean ruda virus, iris potyvirus A, lily yellow mosaic virus, Paris virus 1, and Thunberg fritillary mosaic virus. AcoPV1, AcoPV2, TBV, LMoV, and these nine potyviruses form a robust clade within the genus Potyvirus, supported by a 100% bootstrap value (blue box in Figure 3).
Abundance and distribution of AcoPV1 and AcoPV2 in A. carmichaelii tissues
Potyviruses are primarily transmitted by aphids, which feed on sap from leaves and other easily accessible parts of host plants (Gadhave et al., 2020; Xia et al., 2023). To assess the distribution and abundance of AcoPV1 and AcoPV2 within A. carmichaelii tissue samples, we counted all mapped RNA-seq reads to the viral genome sequences (Table 1).
TABLE 1
| Sample | SRA accession | Totala | AcoPV1b | AcoPV2b |
|---|---|---|---|---|
| Flower | SRR6225420 | 8,643,220 | 18,977 (0.22%) | 75,657 (0.88%) |
| Bud | SRR6225421 | 8,212,299 | 1,680 (0.02%) | 14,782 (0.18%) |
| Leaf | SRR6225422 | 6,301,821 | 44,359 (0.70%) | 22,450 (0.36%) |
| Root | SRR6225423 | 5,662,020 | 2 (<0.01%) | 12 (<0.01%) |
Virus-derived reads in the A. carmichaelii samples.
Total high-quality reads.
Number and percentage of mapped reads.
In flower and leaf tissues, the viral RNA-seq reads of AcoPV1 and AcoPV2 were notably higher compared to bud and root tissues. In the flower tissue, AcoPV1 and AcoPV2 viral reads accounted for 0.22% and 0.88%, respectively, of the total RNA-seq reads. In leaf tissue, viral reads corresponding to the AcoPV1 and AcoPV2 genomes comprised 0.70% and 0.36%, respectively. For bud tissue, we observed lower quantities, with 0.02% for AcoPV1 and 0.18% for AcoPV2. In root tissue, both viruses showed the lowest counts among tissues studied (less than 0.01%).
It is notable that the A. carmichaelii plant analyzed in this study did not display any viral disease symptoms. This suggests that viral infection may be at an initial stage, and their distribution may be limited to the original infection site or nearby tissues. Consequently, the higher abundance of AcoPV1 and AcoPV2 RNA-seq reads in leaf and flower tissues may reflect the feeding preferences of aphids, which are likely their primary vectors.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OP271473, OP271474.
Author contributions
MR, AR, and MY obtained RNA-seq data; DC and YH performed bioinformatics analysis; YH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Government of Korea (grant numbers 2018R1A5A1025077 and RS-2023-00208564).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2023.11782/full#supplementary-material
Footnotes
1.^ https://ictv.global/taxonomy
2.^ https://github.com/najoshi/sickle
3.^ https://github.com/ablab/spades
4.^ https://www.ebi.ac.uk/interpro/download/Pfam
5.^ https://github.com/bbuchfink/diamond
6.^ https://www.ncbi.nlm.nih.gov/orffinder
7.^ https://github.com/wrpearson/fasta36
8.^ http://weblogo.threeplusone.com
9.^ https://github.com/lh3/bwa
10.^ https://mafft.cbrc.jp/alignment/software
References
1
Adams M. J. Antoniw J. F. Beaudoin F. (2005a). Overview and analysis of the polyprotein cleavage sites in the family potyviridae. Mol. Plant Pathol.6, 471–487. 10.1111/j.1364-3703.2005.00296.x
2
Adams M. J. Antoniw J. F. Fauquet C. M. (2005b). Molecular criteria for genus and species discrimination within the family potyviridae. Arch. Virol.150, 459–479. 10.1007/s00705-004-0440-6
3
Bejerman N. Debat H. (2022). Exploring the tymovirales landscape through metatranscriptomics data. Arch. Virol.167, 1785–1803. 10.1007/s00705-022-05493-9
4
Buchfink B. Reuter K. Drost H. G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods18, 366–368. 10.1038/s41592-021-01101-x
5
Bushmanova E. Antipov D. Lapidus A. Prjibelski A. D. (2019). rnaSPAdes: a de novo transcriptome assembler and its application to RNA-seq data. GigaScience8, giz100. 10.1093/gigascience/giz100
6
Capella-Gutierrez S. Silla-Martinez J. M. Gabaldon T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics25, 1972–1973. 10.1093/bioinformatics/btp348
7
Choi D. Shin C. Shirasu K. Ichihashi Y. Hahn Y. (2022). Artemisia capillaris nucleorhabdovirus 1, a novel member of the genus alphanucleorhabdovirus, identified in the artemisia capillaris transcriptome. Acta Virol.66, 149–156. 10.4149/av_2022_204
8
Choi D. Rai M. Rai A. Shin C. Yamazaki M. Hahn Y. (2023). High-throughput RNA sequencing analysis of mallotus japonicus revealed novel polerovirus and amalgavirus. Acta Virol.67, 13–23. 10.4149/av_2023_102
9
Choi D. Shin C. Shirasu K. Hahn Y. (2021). Two novel poty-like viruses identified from the transcriptome data of purple witchweed (Striga hermonthica). Acta Virol.65, 365–372. 10.4149/av_2021_402
10
Chung B. Y. Miller W. A. Atkins J. F. Firth A. E. (2008). An overlapping essential gene in the potyviridae. Proc. Natl. Acad. Sci. U. S. A.105, 5897–5902. 10.1073/pnas.0800468105
11
Crooks G. E. Hon G. Chandonia J. M. Brenner S. E. (2004). WebLogo: a sequence logo generator: figure 1. Genome Res.14, 1188–1190. 10.1101/gr.849004
12
Cruz J. A. Freire A. M. Polimeni J. C. Blawid R. (2023). A novel deltacryptic virus identified in allium cepa from Brazil. Acta Virol.67, 109–113. 10.4149/av_2023_111
13
Gadhave K. R. Gautam S. Rasmussen D. A. Srinivasan R. (2020). Aphid transmission of potyvirus: the largest plant-infecting RNA virus genus. Viruses12, 773. 10.3390/v12070773
14
Goh C. J. Hahn Y. (2021). Analysis of proteolytic processing sites in potyvirus polyproteins revealed differential amino acid preferences of NIa-Pro protease in each of seven cleavage sites. PLoS One16, e0245853. 10.1371/journal.pone.0245853
15
Goh C. J. Park D. Hahn Y. (2021). A novel tepovirus, agave virus T, identified by the analysis of the transcriptome data of blue agave (Agave tequilana). Acta Virol.65, 68–71. 10.4149/av_2021_107
16
Hagiwara-Komoda Y. Choi S. H. Sato M. Atsumi G. Abe J. Fukuda J. et al (2016). Truncated yet functional viral protein produced via RNA polymerase slippage implies underestimated coding capacity of RNA viruses. Sci. Rep.6, 21411. 10.1038/srep21411
17
Ilyas R. Rohde M. J. Richert-Poggeler K. R. Ziebell H. (2022). To be seen or not to be seen: latent infection by tobamoviruses. Plants (Basel)11, 2166. 10.3390/plants11162166
18
Inoue-Nagata A. K. Jordan R. Kreuze J. Li F. Lopez-Moya J. J. Makinen K. et al (2022). ICTV virus taxonomy profile: potyviridae 2022. J. Gen. Virol.103, 001738. 10.1099/jgv.0.001738
19
Kumar S. Stecher G. Li M. Knyaz C. Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35, 1547–1549. 10.1093/molbev/msy096
20
Li H. Durbin R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics25, 1754–1760. 10.1093/bioinformatics/btp324
21
Minh B. Q. Schmidt H. A. Chernomor O. Schrempf D. Woodhams M. D. von Haeseler A. et al (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol.37, 1530–1534. 10.1093/molbev/msaa015
22
Nakamura T. Yamada K. D. Tomii K. Katoh K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics34, 2490–2492. 10.1093/bioinformatics/bty121
23
Nibert M. L. Pyle J. D. Firth A. E. (2016). A +1 ribosomal frameshifting motif prevalent among plant amalgaviruses. Virology498, 201–208. 10.1016/j.virol.2016.07.002
24
Olspert A. Carr J. P. Firth A. E. (2016). Mutational analysis of the potyviridae transcriptional slippage site utilized for expression of the P3N-PIPO and P1N-PISPO proteins. Nucleic Acids Res.44, 7618–7629. 10.1093/nar/gkw441
25
Olspert A. Chung B. Y. Atkins J. F. Carr J. P. Firth A. E. (2015). Transcriptional slippage in the positive-sense RNA virus family potyviridae. EMBO Rep.16, 995–1004. 10.15252/embr.201540509
26
Park D. Goh C. J. Hahn Y. (2021). Two novel closteroviruses, fig virus A and fig virus B, identified by the analysis of the high-throughput RNA-sequencing data of fig (Ficus carica) latex. Acta Virol.65, 42–48. 10.4149/av_2021_104
27
Park D. Kim H. Hahn Y. (2017). Genome sequence of a distinct watermelon mosaic virus identified from ginseng (Panax ginseng) transcriptome. Acta Virol.61, 479–482. 10.4149/av_2017_410
28
Pasin F. Daros J. A. Tzanetakis I. E. (2022). Proteome expansion in the potyviridae evolutionary radiation. FEMS Microbiol. Rev.46, fuac011. 10.1093/femsre/fuac011
29
Rai M. Rai A. Kawano N. Yoshimatsu K. Takahashi H. Suzuki H. et al (2017). De novo RNA sequencing and expression analysis of aconitum carmichaelii to analyze key genes involved in the biosynthesis of diterpene alkaloids. Molecules22, 2155. 10.3390/molecules22122155
30
Rodamilans B. Valli A. Mingot A. San Leon D. Baulcombe D. Lopez-Moya J. J. et al (2015). RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the potyviridae family. J. Virol.89, 6965–6967. 10.1128/JVI.00337-15
31
Rosario K. Van Bogaert N. Lopez-Figueroa N. B. Paliogiannis H. Kerr M. Breitbart M. (2022). Freshwater macrophytes harbor viruses representing all five major phyla of the RNA viral kingdom orthornavirae. PeerJ10, e13875. 10.7717/peerj.13875
32
Schneider T. D. Stephens R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res.18, 6097–6100. 10.1093/nar/18.20.6097
33
Shin C. Choi D. Shirasu K. Hahn Y. (2022a). Identification of dicistro-like viruses in the transcriptome data of striga asiatica and other plants. Acta Virol.66, 157–165. 10.4149/av_2022_205
34
Shin C. Choi D. Shirasu K. Ichihashi Y. Hahn Y. (2022b). A novel RNA virus, thesium chinense closterovirus 1, identified by high-throughput RNA-sequencing of the parasitic plant thesium chinense. Acta Virol.66, 206–215. 10.4149/av_2022_302
35
Untiveros M. Olspert A. Artola K. Firth A. E. Kreuze J. F. Valkonen J. P. (2016). A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing. Mol. Plant Pathol.17, 1111–1123. 10.1111/mpp.12366
36
Wang A. (2021). Cell-to-cell movement of plant viruses via plasmodesmata: a current perspective on potyviruses. Curr. Opin. Virol.48, 10–16. 10.1016/j.coviro.2021.03.002
37
Wen R. H. Hajimorad M. R. (2010). Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology400, 1–7. 10.1016/j.virol.2010.01.022
38
Wylie S. J. Tran T. T. Nguyen D. Q. Koh S. H. Chakraborty A. Xu W. et al (2019). A virome from ornamental flowers in an australian rural town. Arch. Virol.164, 2255–2263. 10.1007/s00705-019-04317-7
39
Xia C. C. Xue W. J. Li Z. Z. Shi J. X. Yu G. F. Zhang Y. (2023). Presenting the secrets: exploring endogenous defense mechanisms in chrysanthemums against aphids. Horticulturae9, 937. 10.3390/horticulturae9080937
40
Yang X. Li Y. Wang A. (2021). Research advances in potyviruses: from the laboratory bench to the field. Annu. Rev. Phytopathol.59, 1–29. 10.1146/annurev-phyto-020620-114550
41
Zheng H. Y. Chen J. Zhao M. F. Lin L. Chen J. P. Antoniw J. F. et al (2003). Occurrence and sequences of lily mottle virus and lily symptomless virus in plants grown from imported bulbs in Zhejiang province, China. Arch. Virol.148, 2419–2428. 10.1007/s00705-003-0207-5
Summary
Keywords
Aconitum potyvirus 1, Aconitum potyvirus 2, polyprotein, PIPO, RNA polymerase slippage
Citation
Choi D, Rai M, Rai A, Yamazaki M and Hahn Y (2023) Discovery of two novel potyvirus genome sequences by high-throughput RNA sequencing in Aconitum carmichaelii tissue samples. Acta Virol. 67:11782. doi: 10.3389/av.2023.11782
Received
08 July 2023
Accepted
03 October 2023
Published
12 October 2023
Volume
67 - 2023
Edited by
Miroslav Glasa, Slovak Academy of Sciences, Slovakia
Reviewed by
Chrisa Orfanidou, Aristotle University of Thessalonik, Greece
David Read, University of Pretoria, South Africa
Updates
Copyright
© 2023 Choi, Rai, Rai, Yamazaki and Hahn.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoonsoo Hahn, hahny@cau.ac.kr
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.