Abstract
Rat hepatitis E virus (RHEV/HEV-C1, species Rocahepevirus ratti) is an emerging zoonotic pathogen, posing an increasing threat to public health worldwide. This study was conducted for better understanding the epidemiology and evolution of RHEV. The isolates sampled so far can be divided into two major genotypes designated a and b. According to the phylogeography, while type a has been detected in four continents, type b is restricted to East and Southeast Asia. Recombination analysis identified three chimeric isolates. Bayesian coalescent analysis suggested that RHEV began to expand around 1956 and was evolving at a high rate. Codon usage bias analysis revealed that RHEV genes are rich in G/C and have additional bias independent of compositional constraints. In codon usage, RHEV is both similar to and different from the major host Norway rat (Rattus norvegicus). Furthermore, unlike many other mammalian RNA viruses, RHEV does not mirror hosts’ marked suppression of “CG” and “TA”.
Introduction
Rat hepatitis E virus (RHEV, species Rocahepevirus ratti), previously known as HEV species C genotype 1 (HEV-C1, species Orthohepevirus C), is the prototype of the genus Rocahepevirus in the family Hepeviridae of RNA viruses (Purdy et al., 2022). It is closely related to ferret hepatitis E virus (FrHEV/HEV-C2) of the same species. Since the first detection in Norway rats (Rattus norvegicus) from Germany (Johne et al., 2010), RHEV variants have been recorded in much of the world (Figure 1), with the Rattus species being the primary hosts. Recently, the population of Rocahepevirus has also expanded rapidly, with a growing number of novel members identified in various rodents from the families Muridae and Cricetidae (Reuter et al., 2020; Wang et al., 2020).
FIGURE 1
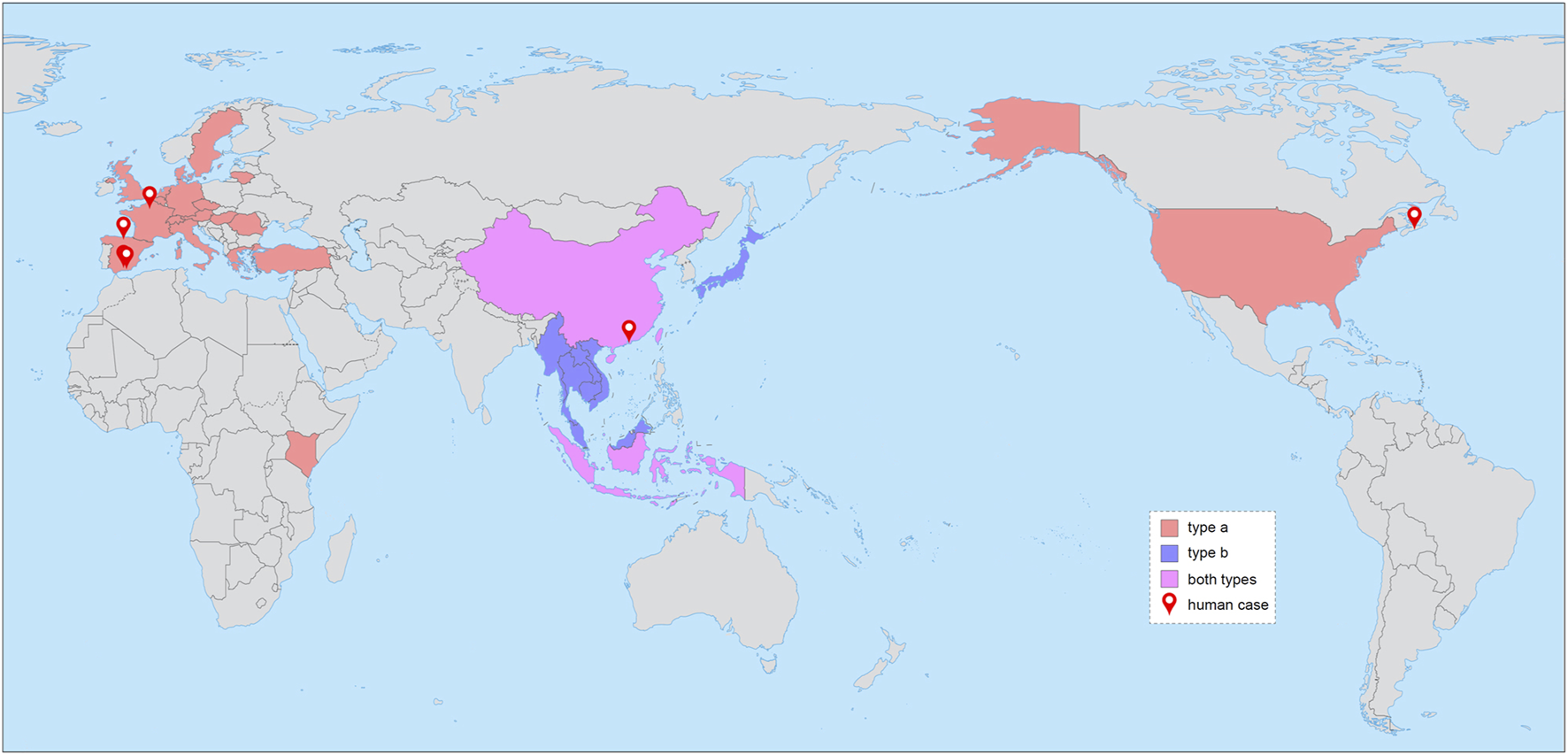
Known geographical distribution of RHEV. The two major genotypes (a and b) are shown in different colors as indicated. The cities where human cases were detected are labeled with map markers. The free world map is from http://bzdt.ch.mnr.gov.cn [No. GS(2016)1665].
Within the subfamily Orthohepevirinae, rocahepeviruses are phylogenetically sister to though highly divergent from paslahepeviruses, which include the well-known HEV (species Paslahepevirus balayani, formerly Orthohepevirus A) that has been imposing a heavy burden on global public health. Four of the eight major genotypes of HEV (1, 2, 3 and 4) are usually responsible for human infection (Primadharsini et al., 2019; Reuter et al., 2020). Until recently, RHEV was demonstrated to be zoonotic like HEV-3 and -4 by a total of 23 patients from China (Sridhar et al., 2018; Sridhar et al., 2021; Sridhar et al., 2022), Canada (Andonov et al., 2019), Spain (Rivero-Juarez et al., 2022), and France (Rodriguez et al., 2023) (Table 1). Notably, immunocompetent individuals are also susceptible, and the clinical features vary greatly, spanning subclinical infection, acute or chronic hepatitis, extrahepatic manifestations, and even fatal outcomes. Moreover, likely spillover events have been observed in several other mammals (Table 1), particularly musk shrews (Suncus murinus), which might represent another viral reservoir (Wang et al., 2020).
TABLE 1
| Species | Country | Case No. | Genotype | First record | |
|---|---|---|---|---|---|
| Year | Accession No. | ||||
| Suncus murinus | China | 13 | a, b | 2012 | LC549186 |
| Ursus arctos syriacus | Germany | 1 | a | 2016 | MF480313 |
| Homo sapiens | China | 16 | b | 2017 | MN450851 |
| Canada | 1 | a | 2017 | MK050105 | |
| Spain | 5 | a | 2018 | OK082153 | |
| France | 1 | a | 2022 | OP610066 | |
| Bos taurus | Turkey | 1 | a | 2018 | MN660075 |
Cross-species events of RHEV.
RHEV has a positive-sense, single-stranded RNA genome of ∼6.9k nucleotides (nt). Between the 5′ 7-methylguanosine cap and the 3′ poly(A) tail, there are four open reading frames (ORFs) (Reuter et al., 2020; Wang et al., 2020) (Figure 2A). The longest ORF1 encodes a nonstructural polyprotein consisting of methyltransferase, papain-like cysteine protease, macro domain, RNA helicase, and RNA-dependent RNA polymerase (RdRp). Adjacent is ORF3 of the multifunctional phosphoprotein. This small ORF largely overlaps with ORF2 of the capsid protein. In addition to ORF4, which lies within ORF1, there are two putative ORFs (5 and 6) in the reference genome (NC_038504/GU345042); however, the two are absent in most of the complete genomes deposited in GenBank.
FIGURE 2
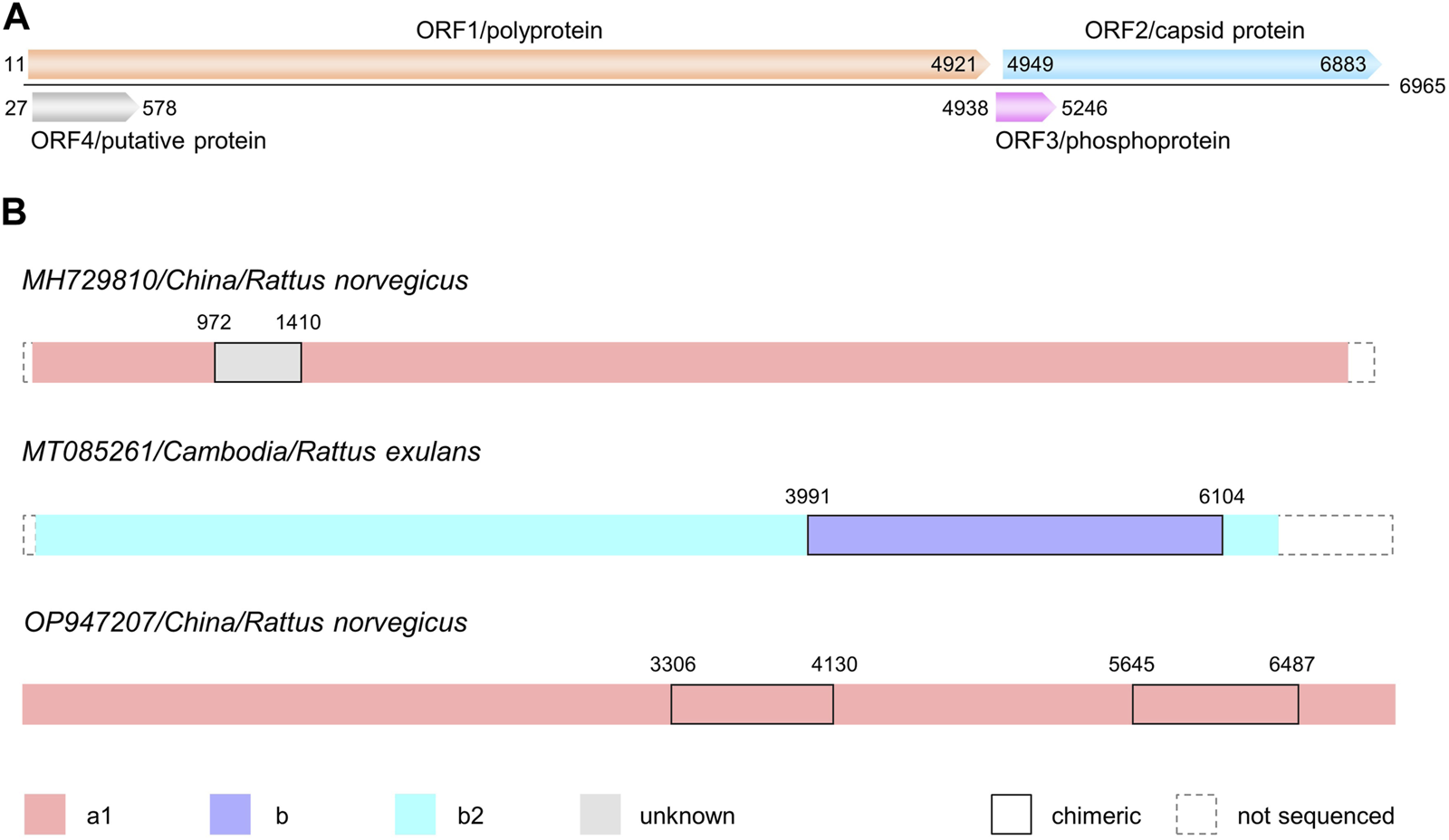
(A) Sketch map of genome organization of RHEV. The four open reading frames (ORFs) are shown in different colors. Position information is from the reference genome NC_038504/GU345042. (B) Chimeric patterns of three RHEV isolates. Segments of different origin are distinguished as indicated. Positions of the putative breakpoints are shown above. The minor parent of MH729810 is currently unknown. a1, b and b2: RHEV genotypes.
Given the ever-growing number of RHEV sequences in GenBank, we aimed to broaden the knowledge on the epidemiology and evolution of this emerging zoonotic pathogen. In this study, the available sequences of RHEV were first submitted to recombination detection. Following the phylogeographical analysis, the Bayesian coalescent approach was applied to a set of time-stamped sequences. In addition, the codon usage bias and dinucleotide composition of RHEV ORFs were measured.
Materials and methods
Available complete and partial sequences of RHEV were downloaded from GenBank (as of August 2023). Multiple sequence alignments were created by using the MUSCLE program executed in MEGA X (Kumar et al., 2018) and then submitted to the RDP package (Takata et al., 2017). All built-in methods including RDP and BootScan were run to seek for possible recombination events. Sequence similarity comparison was visualized by SimPlot (Lole et al., 1999). To show phylogenetic incongruence, the sequences of the chimera and some representative isolates (information given as accession/country/host in Supplementary Figure S1) were segmented according to the suggested breakpoints. Each alignment was then submitted to MEGA X for reconstructing a Maximum Likelihood (ML) tree from 1,000 Bootstrap replicates.
To explain our genotype designation of RHEV, an ML tree was drawn with the partial genomic sequences (trimmed to region 554–6,090, position referred to GU345042) from 47 isolates (accession/country/host in Figure 3). Moreover, most isolates had genomic region 4159–4362 sequenced. Thus, to show the phylogeography and host range of RHEV, another ML tree was drawn based on the 204-nt region (though not sufficient for phylogeny reconstruction) using 131 representative isolates (accession/country/host in Supplementary Figure S2). The trees were generated under the best-fit nucleotide substitution model GTR + G + I determined by MODELTEST in MEGA X.
FIGURE 3
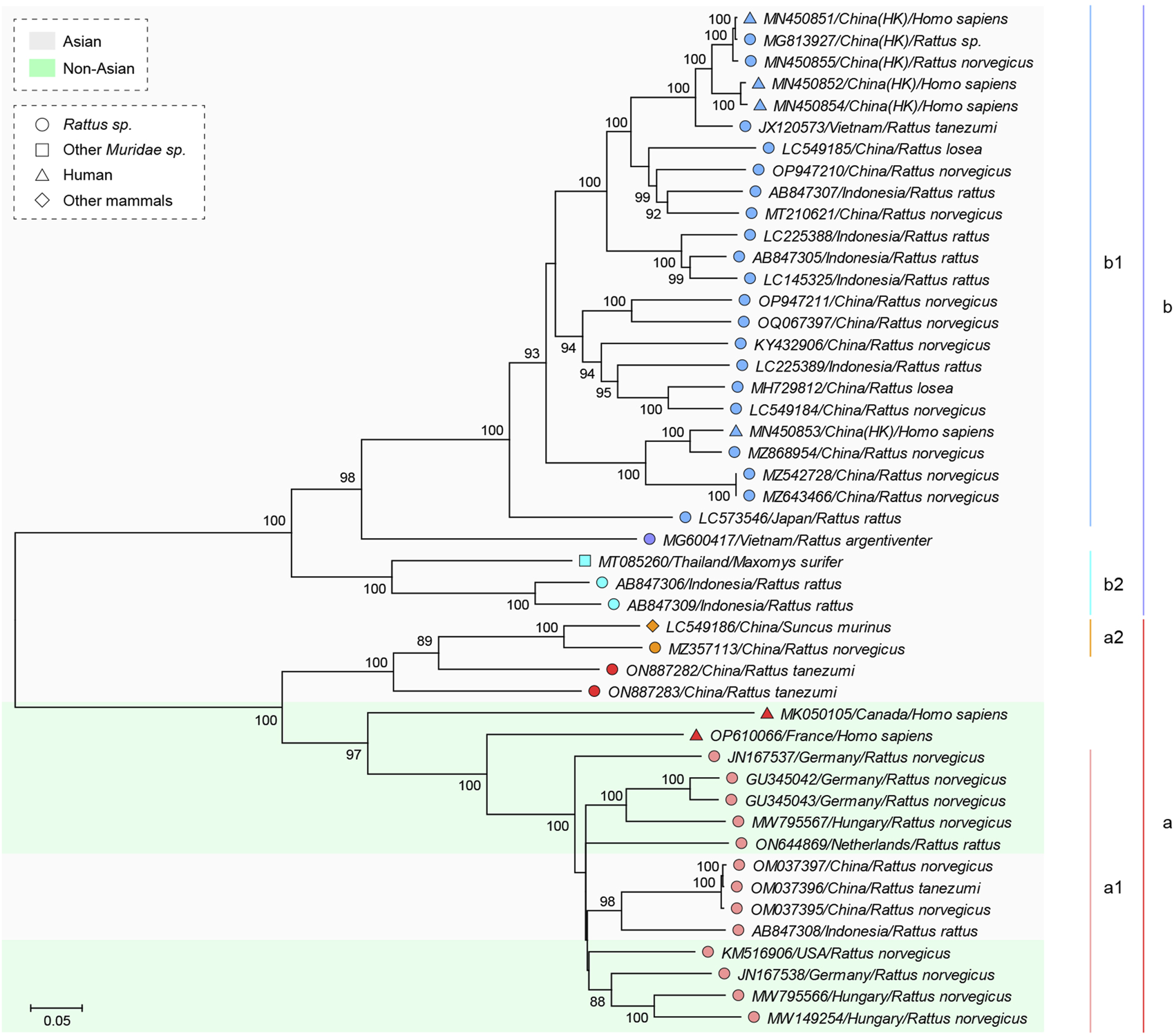
Maximum Likelihood (ML) phylogeny of RHEV based on the genomic region 554–6090. Position information is referred to NC_038504/GU345042. The tree was reconstructed using the sequences of 47 isolates (information given as accession/country/host). Genotype classification is shown on the right. The isolates are differently labeled and shaded as indicated. Branches supported by >70% bootstrap value from 1,000 replicates are indicated.
To date the divergence of RHEV, Bayesian analysis was conducted based on the genomic region 4193–4921 (3′ end of ORF1), given that many time-stamped isolates had the 729-nt region sequenced and a similar region was used for dating the divergence of HEV (Baha et al., 2019). The dataset was composed of 149 unique sequences (accession/country/year/host in Supplementary Figure S3) with collection dates and without ambiguous bases after data cleaning guided by TempEst (Rambaut et al., 2016). The subsets were compiled according to genotype designation. The times to the most recent common ancestor (tMRCAs) and the substitution rates were estimated by the Bayesian Markov chain Monte Carlo (MCMC) method in the BEAST v1.10.4 package (Suchard et al., 2018). Three clock models (strict, exponential, and lognormal) and two demographic models (constant size and exponential growth) were compared for confidence and convergence. The results from independent runs (each 10–25 million MCMC iterations with 10% burn-in) were pulled together to assure convergence (effective sample size > 200). Statistical uncertainty was reflected by the 95% highest probability density (HPD) intervals. The maximum clade credibility (MCC) tree was generated.
To measure the codon usage bias of the four RHEV ORFs, ENC (effective number of codons), GC3S (frequency of G + C at synonymous 3rd codon position), and RSCU (relative synonymous codon usage) values were computed by using CodonW.1 The RSCU values of R. norvegicus genes were calculated based on the reference genome mRatBN7.2 in GenBank. ENC values vary between 20 (extreme bias as one synonymous codon is exclusively used) and 61 (no bias as synonymous codons are evenly used) (Wright, 1990). RSCU value > or < 1.00 stands for more or less frequent usage than expected, respectively. Moreover, to assess the potential effect of host (h) on virus (v) in codon usage, the dissimilarity index was transformed from Pearson Correlation Coefficient (based on the RSCU values of the 59 synonymous codons) using the equation D(v, h) = [1- R(v, h)]/2. The values were thus normalized into the range (0, 1). The higher the value, the larger the difference (Baha et al., 2019).
The dinucleotide composition of RHEV and R. norvegicus ORFs was measured by using DAMBE7 (Xia, 2018). The odds-ratio of observed frequency to expected frequency for dinucleotide XY was calculated as fXY/fXfY (f for nucleotide frequency). The upper and lower value boundaries are 1.23 and 0.78, respectively. An out-of-range value denotes that the dinucleotide is over- or under-represented (Cardon et al., 1994).
Results
Genotype designation and phylogeography of RHEV
For smoother description, the genotype designation of RHEV changed herein was introduced first. The previous designations based on full-length genome were as follows: G1–G3 (Mulyanto et al., 2014), gt1–gt3 (Andonov et al., 2019), and a–d (Bai et al., 2020). Here, according to the ML phylogeny (Figure 3) based on partial genome (region 554–6090), the isolates were divided into two major genotypes (a and b) for convenience. Further, four subtypes (a1, a2, b1, and b2, corresponding to G1/gt1/a, G2/gt3/b, G3/gt2/c, and d, respectively) were designated, excluding five isolates. Such genotype classification was applicable to the ML phylogeny (Supplementary Figure S2) based on the short segment 4159–4362, regardless of the poor robustness.
Then, from the tree in Supplementary Figure S2, which covers all sampling countries except Sweden where two a1-type sequences were detected in wastewater (MW020591-2) (Rivero-Juarez et al., 2022), it is evident that b-type sequences were sampled exclusively from East and Southeast Asia, while a-type sequences were from a much wider area involving four continents. The genotype information was thus included in the world map depicting the known geographical distribution of RHEV (Figure 1). Indonesia and China then appear to be the “hot spot” where both a- and b-type strains are circulating.
Detection of three chimeric sequences of RHEV
When the alignment composed of 44 complete and 12 partial genomic sequences of RHEV was submitted to the RDP package, three significant events were detected. MH729810/China/R. norvegicus (p = 1.296 E-04), MT085261/Cambodia/R. exulans (p = 1.302 E-033), and OP947207/China/R. norvegicus (p = 1.252 E-03), were revealed to be chimeric, as illustrated in Figure 2B.
MH729810 belongs to subtype a1 but for the genomic region 972–1410 (referred to GU345042), where there is a >10% drop in sequence similarity to other isolates. For example, the similarity to AB847308/Indonesia/R. rattus drops from 86.2% to 74.3%. The anomaly is visible when GU345042 serves as the query in the similarity plot (Supplementary Figure S1A). In this region, the decrease of MH729810 contrasts markedly with the increase of AB847308. However, the origin of this region is currently unknown. Given the <75% sequence similarity, the minor parent of MH729810 is likely a novel genotype of RHEV or even the third member of the species Rocahepevirus ratti.
MT085261 is likely derived from intragenotype recombination occurring between the b2 lineage represented by MT085260/Thailand/Maxomys surifer as the major parent and the unassigned b lineage represented by MT085262/Cambodia/R. exulans as the minor parent. It is chimeric in the genomic region 3991–6104. Such pattern is demonstrated by the phylogenetic incongruence of different regions partitioned according to the putative breakpoints. As is clear in Supplementary Figure S1B, MT085261 is clustered within the clade of either b2 or unassigned b, depending on the region analyzed. Nevertheless, considering that the three isolates were sequenced in the same lab, it cannot be asserted that the recombination event occurred naturally.
OP947207 was detected by RDP to be chimeric in the genomic region 3306–4130, with OP947209/China/Suncus murinus and OP947206/China/Mus musculus being the putative major and minor parents, respectively. In the similarity plot (Supplementary Figure S1C), OP947207 does exhibit a drop (99.2%–91.8%) in sequence similarity to OP947209 in this region. Interestingly, there is a second drop (99.2%–92.8%) in the genomic region 5645–6487. Judging from the wavy line of OP947206, however, there is no particular increase in either region. In fact, OP947207 only has a relatively higher sequence similarity to OP947206 in the first region (92.8% vs. 91.8%). Moreover, even OP947206 and OP947209 share 91.8% sequence similarity, clustering together in subtype a1 (Supplementary Figure S2). It is thus more likely that the unusual pattern of OP947207 resulted from divergent evolution (away from OP947209) occurring in the two regions.
Dated divergence of RHEV
The dataset composed of 149 unique sequences (genomic region 4193–4921) collected between 2007 and 2020 (Supplementary Figure S3) was submitted to Bayesian coalescent analysis. The best-fit substitution model was also GTR + G + I. The relaxed uncorrelated exponential clock could be used, whereas the other two clock models (strict and lognormal) failed to reach convergence. The constant growth model was selected for being usable for all datasets.
Then, the tMRCAs were estimated for the RHEV genotypes, as listed in Table 2. The MRCA of the 149 isolates emerged 64 (95% HPD: 35–102) years before 2020. In other words, the first bifurcation event generating the two major genotypes occurred around 1956 (1918–1985), as shown in the time-scaled MCC tree (Supplementary Figure S3). Later, the MRCA of the 68 a-type isolates emerged around 1977 (1955–1995), 1 year before that of the 81 b-type isolates. Similarly, the MRCA of the 66 a1-type isolates emerged around 1995 (1986–2002), 1 year before that of the 78 b1-type or three b2-type isolates. The latest one was the MRCA of the two a2-type isolates that emerged around 2010 (2007–2012).
TABLE 2
| Genotype | No. of sequences | tMRCAa | Age | Rate (×10–2) | |||
|---|---|---|---|---|---|---|---|
| Mean | 95% HPD | Mean | 95% HPD | Mean | 95% HPD | ||
| all | 149 | 64 | 35–102 | 1956 | 1918–1985 | 1.06 | 0.74–1.41 |
| a | 68 | 43 | 25–65 | 1977 | 1955–1995 | 1.63 | 0.81–2.53 |
| b | 81 | 42 | 25–64 | 1978 | 1956–1995 | 0.93 | 0.43–1.41 |
| a1 | 66 | 25 | 18–34 | 1995 | 1986–2002 | 1.66 | 0.98–2.36 |
| a2 | 2 | 10 | 8–13 | 2010 | 2007–2012 | — | |
| b1 | 78 | 24 | 16–34 | 1996 | 1986–2004 | 1.07 | 0.57–1.60 |
| b2 | 3 | 24 | 15–36 | 1996 | 1984–2005 | — | |
Bayesian estimates of RHEV genotypes.
tMRCA, time to the most recent common ancestor (years before 2020); HPD, highest probability density.
The average nucleotide substitution rate of the 149 isolates was calculated to be 1.06 (0.74–1.41) × 10−2 subs/site/year. Additionally, rates were calculated for the genotypes (Table 2) using the corresponding subsets under the same Bayesian parameters. The rate of type a was even higher at 1.63 (0.81–2.53) × 10−2, while that of type b was lower at 0.93 (0.43–1.41) × 10−2. With further division into subtypes a1 and b1, the rates increased to 1.66 (0.98–2.36) × 10−2 and 1.07 (0.57–1.60) × 10−2, respectively. Notably, the relaxed uncorrelated lognormal clock was also able to describe the evolutionary dynamics of the 66 a1-type isolates and yielded similar results with an age of 1996 (1984–2004) and a rate of 1.86 (1.14–2.63) × 10−2.
Codon usage bias and dinucleotide composition of RHEV ORFs
After calculation, ENC was plotted against GC3S to visually display synonymous codon usage bias of RHEV ORFs. As shown in Figure 4A, the ENC values of all points except some of ORF3 exceed 40, which is indicative of weak bias. All ORF3 points, regardless of the genotype, are distributed away from the other points, showing generally lower ENC values (avg. 39.87) and markedly higher GC3S values (avg. 0.726). In fact, the codon choice of ORF3 is restricted not only by the (3, 1) overlapping pattern (the 3rd codon position of ORF3 is the 1st codon position of ORF2) but by the short gene length (∼309 nt).
FIGURE 4
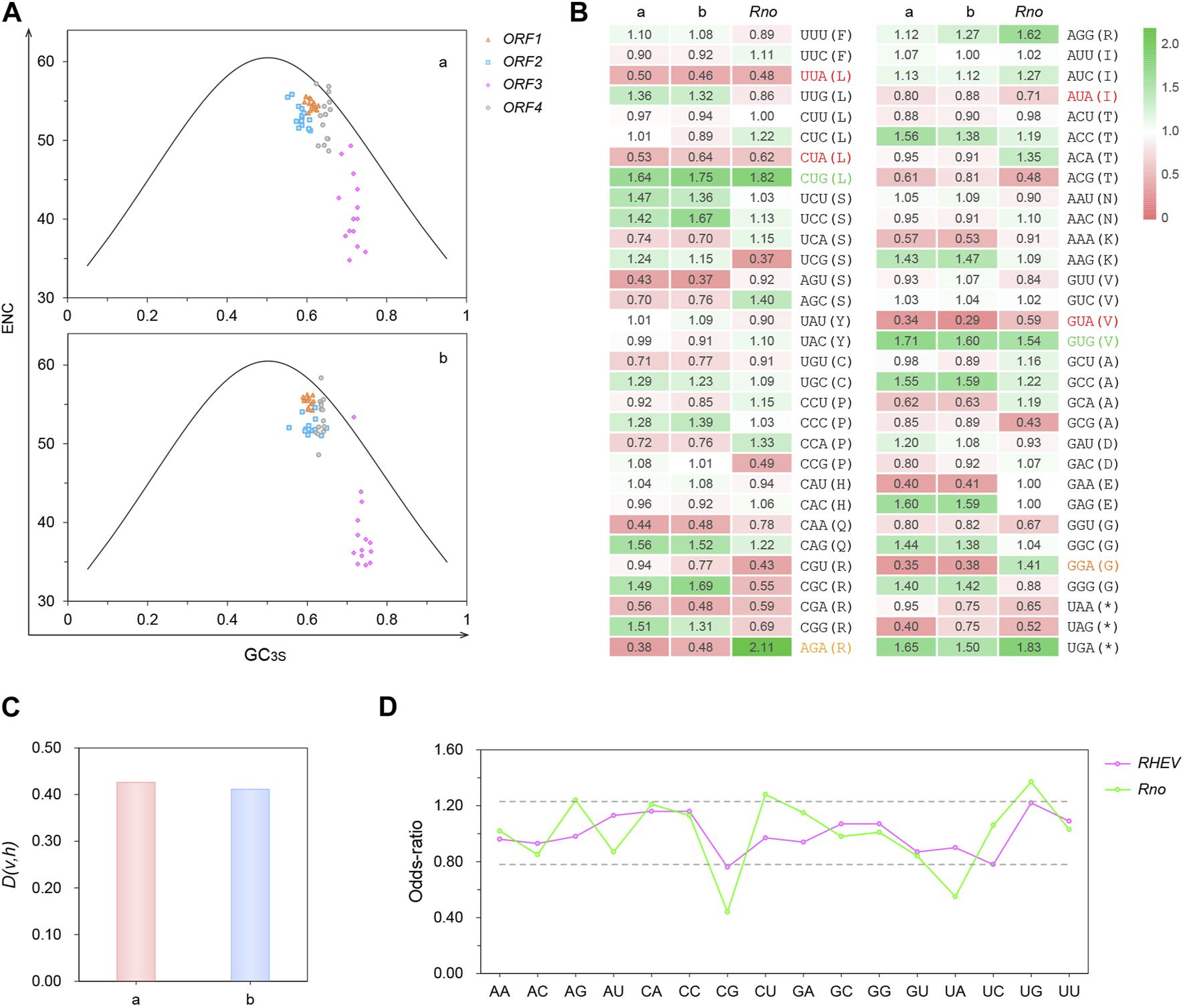
(A) ENC-plots. ENC (effective number of codons) is plotted against GC3S (frequency of G + C at synonymous 3rd codon position) for the four RHEV ORFs (open reading frames) in different forms and colors. The solid curve is formed by the theoretical values shaped only by GC compositional constraints. The plots were drawn separately for the two major genotypes (a and b). (B) Heatmap of the overall RSCU values of RHEV and Norway rat (Rattus norvegicus) genes. RSCU (relative synonymous codon usage) value > or < 1.00 denotes more or less frequent usage than expected, respectively. Codons in red are disfavored by both virus and host, in contrast to those in green. Codons in orange are confronted with opposite bias. (C) Dissimilarity between RHEV and R. norvegicus in codon usage. D(v, h): dissimilarity index (the higher the number, the larger the difference). (D) Dinucleotide composition in the ORFs of RHEV and R. norvegicus. The upper and lower value boundaries shown in dashed lines are 1.23 and 0.78, respectively. An out-of-range value denotes that the dinucleotide is over- or under-represented.
In the ENC-plots (Figure 4A), the actual ENC values are plotted alongside the curve formed by the theoretical ENC values. A point lying on or just below the curve, as exemplified by the ORF4 point of MK050105/Canada/Homo sapiens, denotes that the gene is subject to GC compositional constraints, from mutation pressure biased toward G/C or translational selection for codons ending in G/C (Wright, 1990). For the points lying away from the curve, their codon choice is under other selection pressure. One possible influence is protein aromaticity, which is in significant correlation (R = 0.85, p < 0.001) with ENC.
From the overall RSCU values of RHEV genes (Figure 4B), a preference of “G” over “A” was observed in most synonymous 3rd codon positions, which is in accordance with the high GC3S values (Figure 4A, avg. 0.643). Indeed, as confirmed by the results of nucleotide composition in the synonymous 3rd codon position (Supplementary Figure S4A), A3S is much lower than G3S in all four ORFs (0.139 vs. 0.304). Nevertheless, it is “C” that is more over-represented (0.338), whereas “U” is under-represented in ORF3 (0.152) and ORF4 (0.190). Like the HEV counterparts (Baha et al., 2019), all RHEV ORFs, particularly ORF3, are rich in G/C, with the average GC content reaching 0.597 (Supplementary Figure S4A).
The neutrality plot (Sueoka, 1988), in which GC12S was plotted against GC3S, was then drawn to quantify the mutation-selection equilibrium in shaping the overall codon usage bias of RHEV ORFs. As shown in Supplementary Figure S4B, there is a significant positive correlation between GC12S and GC3S (R = 0.80, p < 0.001). The slope of the regression line is 0.74, indicating that the influences of mutation pressure and natural selection are 74% and 26%, respectively; that is, mutation pressure is the dominant force. Indeed, ENC is in significant correlation with GC (R = −0.89, p < 0.001).
When compared with the major host R. norvegicus (Figure 4B), a notable similarity in the overall RSCU values is that both species have a bias against the codons ending in “UA”. This is likely due to host’s “UA” deficiency discussed below. In contrast, both species show a high preference for “CUG” and “GUG”. Nevertheless, RHEV does not fully follow the codon preference of R. norvegicus. For example, “AGA” and “GGA” are highly favored by the host but disfavored by the virus. Such difference is reflected by the high D(v, h) value (0.419) (note that the dissimilarity between R. norvegicus and H. sapiens is valued at only 0.004). Further, the two major genotypes were compared for host similarity. As shown in Figure 4C, type a is more different from the host than type b (0.426 vs. 0.411), suggesting that the codon usage of type a is less affected by host shaping. In addition, the codon usage bias of RHEV is largely similar to that of HEV (Baha et al., 2019). Particularly, both viruses have a strong bias toward the sense codons ending in “AG” but against those ending in “UA”, “GA” and “AA”.
From the RSCU values (Figure 4B), another feature noticed is that the dinucleotide “CG” is particularly avoided in the codons by the host but not the virus. Indeed, as revealed by the dinucleotide composition results (Figure 4D), “CG” is only slightly under-represented (<0.78) in RHEV ORFs (0.76), in contrast to the marked deficiency in R. norvegicus ORFs (0.44). Surprisingly, although “UA” is particularly avoided at the (2, 3) codon positions in both species, the virus is not deficient in “UA” (0.90), unlike the host (0.55).
Discussion
According to the phylogeography of RHEV (Supplementary Figure S2), while type b is restricted to East and Southeast Asia, type a has been detected in four continents (Figure 1). Notably, the Canadian patient was supposed to catch the infection during the stay in the Democratic Republic of Congo and Gabon (Andonov et al., 2019). Meanwhile, an a1-type sequence (MK935162) was sampled from R. rattus in Kenya (Onyuok et al., 2019). It is thus possible that RHEV is endemic in central Africa. Moreover, RHEV may be endemic in India and the USA, as suggested by the high seroprevalence rates of HEV antibodies in some Muridae species (Wang et al., 2020) [note that it is unknown whether the French patient was infected in Europe or India (Rodriguez et al., 2023)]. Due to the lack of sequence information in these regions, the origin of RHEV cannot be safely inferred from the current phylogeography. Nevertheless, there is little doubt that Southeast Asia is the cradle of the b lineage (Figure 3 and Supplementary Figure S2), though sampling bias is also present.
From the host information included in Figure 3 or Supplementary Figure S2, it can be derived that both a and b types can infect humans and shrews (listed in Table 1). Perhaps, RHEV is naturally infectious to most mammals and cross-species transmission is more dependent on the odds of close contact with the infected rats. Notably, all Rocahepevirus members except FrHEV are basically harbored by rodents. They can be split into two groups that infect Muridae and Cricetidae species, respectively, according to the ML phylogeny by Reuter et al. (2020). Given that FrHEV/HEV-C2 falls into the Muridae-infecting group, sharing with RHEV/HEV-C1 a common ancestor sister to HEV-C3 (tentative), it is highly possible that FrHEV represents an old host switch event from Muridae of Rodentia to Mustelidae of Carnivora. There is a special case that the isolate KU670940 from a common kestrel (Falco tinnunculus) is clustered with the rocahepeviruses from common voles (Microtus arvalis) (MK192405-9), but not the avihepeviruses (formerly Orthohepevirus B) from birds. It appears that hunting renders rocahepeviruses chances for host jumping from prey to predator.
In recombination analysis, three chimeric RHEV isolates (Figure 2B) were identified. It is not surprising that RHEV can undergo recombination to drive population variability, since several recombinants have been identified for HEV (Smith et al., 2020), one of which is even derived from a double event involving both intra- and inter-genotype recombination (Wang et al., 2010). In fact, it has been shown that the entire Hepeviridae family likely arose from an ancient recombination event occurring between plant and insect viruses. The breakpoint was located at the junction of the nonstructural and structural encoding regions, leading to an “alpha-like” ORF1 and a “Picorna-like” ORF2 (Kelly et al., 2016). Notably, the potential intergenotype recombination event of MH729810 also warns of rapid emergence of a distinct viral strain or species with possibly dangerous consequences.
According to Bayesian estimates (Table 2), the MRCA of the 149 RHEV isolates emerged just around 1956. Compared with HEV, RHEV has a much younger MRCA, which is even younger than the MRCA of HEV-1 (emerging around a century ago), the youngest one among the four human-infecting HEV genotypes (Purdy and Khudyakov, 2010; Baha et al., 2019). The average rate of RHEV was 1.06 × 10−2 subs/site/year. This is quite high, although falling into the range of evolutionary rates documented for RNA viruses (10–5 to 10–1) (Sanjuan, 2012). It is similar to the rates of poliovirus 1 (PV1) (1.17 × 10−2) and PV2 (1.01 × 10−2), two members of the species Enterovirus C in the family Picornaviridae, but higher than the rates of most RNA viruses. In particular, it is ∼6-fold higher than the rate estimated for HEV (1.76 × 10−3) based on an 852-nt segment at the 3′ end of ORF1 under the same Bayesian models (Baha et al., 2019). It is not impossible that the RdRp of RHEV is more error-prone than that of HEV, since the two are highly divergent from each other. Although it is unknown whether the variance is associated with host difference, such high speed of evolution adds fuel to the zoonotic threat of RHEV.
In codon usage, RHEV is both similar to and different from R. norvegicus (Figure 4B). Such information might be useful in manipulating viral gene expression and designing attenuated viruses (Haas et al., 1996). It might be able to lower viral gene expression and virulence via deoptimizing the synonymous codons with those disfavored by both virus and host [e.g., “UUA” (L), “CGA” (R) and “GUA” (V)] and/or decreasing the number of the codons favored by both species [e.g., “CUG” (L) and “CAG” (Q)]. However, given the particular disfavor, increasing the number of “AGA” (R) and “GGA” (G), which are abundant codons in the host, might have a negative effect on viral viability rather than elevating viral gene expression. Notably, these might be extended to HEV, since the usage patterns of the mentioned codons are similar (Baha et al., 2019).
In mammalian genomes, “CG” and “TA” are markedly under-represented, which may result from DNA deamination following methylation and selection for increased mRNA stability, respectively (Simmonds et al., 2013). Such compositional abnormalities appear to be drawn on in cellular antiviral defense. In particular, zinc-finger antiviral protein (ZAP) has been identified as a powerful restriction factor active in “CG” and “UA” surveillance against non-self RNAs. Upon detection, ZAP can directly bind to the targeted sequences, leading to suppression of viral replication (Takata et al., 2017; Odon et al., 2019).
Unlike many other mammalian RNA viruses, including hepatitis A virus (HAV, species Hepatovirus A, family Picornaviridae) but not hepatitis C virus (HCV, species Hepacivirus hominis, family Flaviviridae) (Simmonds et al., 2013; Di Giallonardo et al., 2017), RHEV does not mirror hosts’ marked suppression of “CG” and “TA” (Figure 4D). Nevertheless, in RHEV ORFs, “UA” is particularly avoided at the (2, 3) codon positions (Figure 4B). Then, tRNA abundance is the likely factor accounting for the bias against the codons ending in “UA”, rather than ribonucleases such as RNAseL that can target “UA” for RNA degradation (Odon et al., 2019).
In general, mammalian hepeviruses are not particularly biased against “CG” and “UA”, nor are the closest relative viruses including rubella virus (RuBV, species Rubivirus rubellae, family Matonaviridae) and togaviruses (Rima and McFerran, 1997; Di Giallonardo et al., 2017). Perhaps, these viruses have developed an evasive/resistant strategy against “CG” and “UA” surveillance by ZAP and other antiviral factors, or even exploited the restriction to fine-tune the replication to maximize evolutionary fitness. Then, artificially increasing “CG” and/or “UA” frequencies, which is able to attenuate various RNA viruses (Odon et al., 2019), is not suitable for these viruses.
In summary, three RHEV sequences were identified as chimeric. Bayesian coalescent analysis with the time-stamped genomic sequences suggested that RHEV began to expand in the mid-20th century and was evolving at a very high rate. RHEV ORFs are rich in G/C and have additional bias independent of compositional constraints. In codon usage, RHEV is both similar to and different from the major host R. norvegicus. Moreover, RHEV does not mirror hosts’ marked suppression of “CG” and “TA”.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2023.12031/full#supplementary-material
Footnotes
References
1
Andonov A. Robbins M. Borlang J. Cao J. Hatchette T. Stueck A. et al (2019). Rat hepatitis E virus linked to severe acute hepatitis in an immunocompetent patient. J. Infect. Dis.220, 951–955. 10.1093/infdis/jiz025
2
Baha S. Behloul N. Liu Z. Wei W. Shi R. Meng J. (2019). Comprehensive analysis of genetic and evolutionary features of the hepatitis E virus. BMC Genomics20, 790. 10.1186/s12864-019-6100-8
3
Bai H. Li W. Guan D. Su J. Ke C. Ami Y. et al (2020). Characterization of a novel rat hepatitis E virus isolated from an Asian musk shrew (Suncus murinus). Viruses12, 715. 10.3390/v12070715
4
Cardon L. R. Burge C. Clayton D. A. Karlin S. (1994). Pervasive CpG suppression in animal mitochondrial genomes. Proc. Natl. Acad. Sci. U. S. A.91, 3799–3803. 10.1073/pnas.91.9.3799
5
Di Giallonardo F. Schlub T. E. Shi M. Holmes E. C. (2017). Dinucleotide composition in animal RNA viruses is shaped more by virus family than by host species. J. Virol.91, e02381. 10.1128/JVI.02381-16
6
Haas J. Park E. C. Seed B. (1996). Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol.6 (3), 315–324. 10.1016/s0960-9822(02)00482-7
7
Johne R. Plenge-Bonig A. Hess M. Ulrich R. G. Reetz J. Schielke A. (2010). Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol.91, 750–758. 10.1099/vir.0.016584-0
8
Kelly A. G. Netzler N. E. White P. A. (2016). Ancient recombination events and the origins of hepatitis E virus. BMC Evol. Biol.16, 210. 10.1186/s12862-016-0785-y
9
Kumar S. Stecher G. Li M. Knyaz C. Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35, 1547–1549. 10.1093/molbev/msy096
10
Lole K. S. Bollinger R. C. Paranjape R. S. Gadkari D. Kulkarni S. S. Novak N. G. et al (1999). Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol.73, 152–160. 10.1128/JVI.73.1.152-160.1999
11
Mulyanto S. Andayani J. B. Khalid I. G. Takahashi M. Ohnishi H. Jirintai S. et al (2014). Marked genomic heterogeneity of rat hepatitis E virus strains in Indonesia demonstrated on a full-length genome analysis. Virus Res.179, 102–112. 10.1016/j.virusres.2013.10.029
12
Odon V. Fros J. J. Goonawardane N. Dietrich I. Ibrahim A. Alshaikhahmed K. et al (2019). The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res.47, 8061–8083. 10.1093/nar/gkz581
13
Onyuok S. O. Hu B. Li B. Fan Y. Kering K. Ochola G. O. et al (2019). Molecular detection and genetic characterization of novel RNA viruses in wild and synanthropic rodents and shrews in Kenya. Front. Microbiol.10, 2696. 10.3389/fmicb.2019.02696
14
Primadharsini P. P. Nagashima S. Okamoto H. (2019). Genetic variability and evolution of hepatitis E virus. Viruses11, 456. 10.3390/v11050456
15
Purdy M. A. Drexler J. F. Meng X. J. Norder H. Okamoto H. Van der Poel W. H. M. et al (2022). ICTV virus taxonomy profile: Hepeviridae 2022. J. Gen. Virol.103. 10.1099/jgv.0.001778
16
Purdy M. A. Khudyakov Y. E. (2010). Evolutionary history and population dynamics of hepatitis E virus. PLoS One5, e14376. 10.1371/journal.pone.0014376
17
Rambaut A. Lam T. T. Max Carvalho L. Pybus O. G. (2016). Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol.2, vew007. 10.1093/ve/vew007
18
Reuter G. Boros A. Pankovics P. (2020). Review of hepatitis E virus in rats: evident risk of species Orthohepevirus C to human zoonotic infection and disease. Viruses12, 1148. 10.3390/v12101148
19
Rima B. K. McFerran N. V. (1997). Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J. Gen. Virol.78 (Pt 11), 2859–2870. 10.1099/0022-1317-78-11-2859
20
Rivero-Juarez A. Frias M. Perez A. B. Pineda J. A. Reina G. Fuentes-Lopez A. et al (2022). Orthohepevirus C infection as an emerging cause of acute hepatitis in Spain: first report in Europe. J. Hepatol.77, 326–331. 10.1016/j.jhep.2022.01.028
21
Rodriguez C. Marchand S. Sessa A. Cappy P. Pawlotsky J. M. (2023). Orthohepevirus C hepatitis, an underdiagnosed disease?J. Hepatol.79, e39–e41. 10.1016/j.jhep.2023.02.008
22
Sanjuan R. (2012). From molecular genetics to phylodynamics: evolutionary relevance of mutation rates across viruses. PLoS Pathog.8, e1002685. 10.1371/journal.ppat.1002685
23
Simmonds P. Xia W. Baillie J. K. McKinnon K. (2013). Modelling mutational and selection pressures on dinucleotides in eukaryotic phyla--selection against CpG and UpA in cytoplasmically expressed RNA and in RNA viruses. BMC Genomics14, 610. 10.1186/1471-2164-14-610
24
Smith D. B. Izopet J. Nicot F. Simmonds P. Jameel S. Meng X. J. et al (2020). Update: proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A). J. Gen. Virol.101, 692–698. 10.1099/jgv.0.001435
25
Sridhar S. Yip C. C. Y. Lo K. H. Y. Wu S. Situ J. Chew N. F. S. et al (2022). Hepatitis E virus species C infection in humans, Hong Kong. Clin. Infect. Dis.75, 288–296. 10.1093/cid/ciab919
26
Sridhar S. Yip C. C. Y. Wu S. Cai J. Zhang A. J. Leung K. H. et al (2018). Rat hepatitis E virus as cause of persistent hepatitis after liver transplant. Emerg. Infect. Dis.24, 2241–2250. 10.3201/eid2412.180937
27
Sridhar S. Yip C. C. Y. Wu S. Chew N. F. Leung K. H. Chan J. F. et al (2021). Transmission of rat hepatitis E virus infection to humans in Hong Kong: a clinical and epidemiological analysis. Hepatology73, 10–22. 10.1002/hep.31138
28
Suchard M. A. Lemey P. Baele G. Ayres D. L. Drummond A. J. Rambaut A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol.4, vey016. 10.1093/ve/vey016
29
Sueoka N. (1988). Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. U. S. A.85, 2653–2657. 10.1073/pnas.85.8.2653
30
Takata M. A. Goncalves-Carneiro D. Zang T. M. Soll S. J. York A. Blanco-Melo D. et al (2017). CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature550, 124–127. 10.1038/nature24039
31
Wang B. Harms D. Yang X. L. Bock C. T. (2020). Orthohepevirus C: an expanding species of emerging hepatitis E virus variants. Pathogens9, 154. 10.3390/pathogens9030154
32
Wang H. Zhang W. Ni B. Shen H. Song Y. Wang X. et al (2010). Recombination analysis reveals a double recombination event in hepatitis E virus. Virol. J.7, 129. 10.1186/1743-422X-7-129
33
Wright F. (1990). The 'effective number of codons' used in a gene. Gene87, 23–29. 10.1016/0378-1119(90)90491-9
34
Xia X. (2018). DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol.35, 1550–1552. 10.1093/molbev/msy073
Summary
Keywords
rat hepatitis E virus, recombination, divergence, codon usage, dinucleotide
Citation
Zhao L and Huang Y (2023) Evolution of rat hepatitis E virus: recombination, divergence and codon usage bias. Acta Virol. 67:12031. doi: 10.3389/av.2023.12031
Received
11 September 2023
Accepted
15 November 2023
Published
27 November 2023
Volume
67 - 2023
Edited by
Boris Klempa, Slovak Academy of Sciences, Slovakia
Reviewed by
Sunil Kumar Dubey, Columbia University, United States
Assen Marintchev, Boston University, United States
Ľuboš Korytár, University of Veterinary Medicine and Pharmacy in Košice, Slovakia
Ayalew Mergia, University of Florida, United States
Updates
Copyright
© 2023 Zhao and Huang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangmei Huang, ymhuangcq@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.