- 1Department of Microbiology and Virology, Kerman University of Medical Sciences, Kerman, Iran
- 2Nuclear Agriculture Research School, Nuclear Science and Technology Research Institute (NSTRI), Karaj, Iran
- 3Australian Red Cross Lifeblood, West Melbourne, VIC, Australia
- 4Department of Pharmacognosy and Pharmaceutical Biotechnology, School of Pharmacy, Iran University of Medical Sciences, Tehran, Iran
- 5Department of Computer Science, Modeling, Electronics and Systems Engineering (DIMES), University of Calabria, Rende, Province of Cosenza, Italy
- 6Department of Surgical and Medical Sciences, Magna Graecia University of Catanzaro, Catanzaro, Italy
Long non-coding RNAs (lncRNAs) have gained prominence due to their involvement in various cellular processes, but their specific roles remain elusive. Dysregulation of lncRNAs has been implicated in the pathogenesis of several diseases. In this study, we aimed to shed light on the role of lncRNAs in individuals infected with human immunodeficiency virus type 1 (HIV-1) by examining their changes in the expression patterns related to the initiation of antiretroviral therapy (ART) during acute or chronic phases of infection, compared to healthy controls. We found 316 differentially expressed (DE) lncRNAs in patients receiving long-term ART, shedding light on their potential roles. We also observed interactions between these DE lncRNAs and specific microRNAs (miRNAs). Some of these miRNAs, such as hsa-miR-574-5p, hsa-miR-765, hsa-miR-6165, hsa-miR-1207-5p, and hsa-miR-378i, are associated with cancer progression or suppression, while others, including hsa-miR-328-5p, hsa-miR-4753-3p, and MiR-664, play roles in immune system regulation. Furthermore, our study revealed substantial enrichment in distinct Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, such as axon guidance, PI3K-Akt signaling, and MAPK signaling pathways. Although our results indicate possible molecular processes impacted by the discovered lncRNAs, we cannot explicitly establish causality or specific connections between lncRNAs and genes in these pathways, fostering more specific studies. Furthermore, Gene Ontology (GO) analysis highlighted terms such as cytoskeletal protein binding, ion channel function, synaptic processes, neuron projection, and the somatodendritic compartment, underscoring the relevance of lncRNAs in these cellular components within the context of HIV-1 infection and ART treatment. In conclusion, our study emphasizes the need for further exploration of lncRNAs as potential biomarkers and therapeutic targets in HIV-1-infected patients, with a particular focus on CD4+ T cells. Understanding the functions of lncRNAs in these contexts may pave the way for novel treatment strategies and improved patient outcomes, aligning with the broader goals of our research.
Introduction
Acquired immunodeficiency syndrome (AIDS), a disease caused by human immunodeficiency virus type 1 (HIV-1), is responsible for millions of deaths worldwide (De Boer et al., 2003; Margolis et al., 2020). The disease results in the susceptibility of the body to opportunistic infections. This occurs because of immunodeficiency caused by the death of CD4+ T cells infected with the HIV-1 virus (Campos Coelho et al., 2020). The current common treatment for HIV-1 consists of drugs that target viral enzymes, such as nucleoside and non-nucleoside reverse transcriptase inhibitors (NRTIs and NNRTIs) and protease and integrase strand transfer inhibitors (PIs and INSTIs) (Bandera et al., 2019). Studies in infected humans have shown that antiretroviral therapy (ART) virtually reduces plasmatic viremia by stopping viral replication in the blood (Bale and Kearney, 2019; Bandera et al., 2019; Margolis et al., 2020). In the absence of ART, viral genome integration into host cell DNA occurs, leading to productive infection and the rapid death of CD4+ T cells (Leyre et al., 2020).
HIV-1 infection cannot be completely cured by ART, and discontinuing treatment at any time in a patient’s life would lead to reactivation of the virus because HIV-1 mainly uses CD4+ memory T cells as a reservoir. The fact that these cells have a long half-life (nearly 500 days) helps the virus maintain latency in the host for life (De Boer et al., 2003; Chao et al., 2019; Campos Coelho et al., 2020). The timing of the onset of ART and the immunologic status of the individual are two of the most important factors in the risk of death in patients with HIV-1 infection (Chopra et al., 2019). In 2015, the World Health Organization (WHO) amended the criteria for introducing antiretroviral therapy (ART). These guidelines now propose that all people infected with HIV-1 should begin ART at the time of their HIV-1 diagnosis and potentially even during the acute infection phase. The primary objective of this recommendation is to prevent the deterioration of the immune system and the inflammation caused by the virus (Petkov and Chiodi, 2022). However, further studies are needed to determine how the mentioned factors affect the outcome of HIV-1 infection (Morcilla et al., 2021).
In recent years, scientists have been able to study eukaryotic genomes in greater detail. This has been made possible by the advantages of next-generation sequencing, clustered regularly interspaced short palindromic repeats gene editing, RNA deep sequencing (RNA-Seq), and other developments in high-throughput gene sequencing technologies (Ørom et al., 2010; Moran et al., 2012; Chen and Shan, 2020; Zou and Xu, 2020). The results of studies have highlighted that only a small fraction of the human genome (<2%) is responsible for protein coding, and the rest consists of non-coding genes that produce non-coding RNA transcripts (ncRNAs) (Ørom et al., 2010; Moran et al., 2012; Zou and Xu, 2020). LncRNAs are transcripts (>200 nts) that lack protein-coding RNA. It has been demonstrated that lncRNAs play a critical role in the regulation of various cellular pathways, including viral diseases such as HIV-1 (Boliar et al., 2019).
An important functional area in which lncRNAs may have an impact is the host response to infectious particles such as viruses by regulating host responses and participating in the primary immune response (Peng et al., 2010; Wang et al., 2019). Research on lncRNAs could be of great benefit as they can alter disease progression, infection, and host response. Studying the function of lncRNAs may provide us with new biological indicators to track the progression of various diseases. Identifying new lncRNAs that are differentially expressed as a cause of viral infection may bring us one step closer to understanding their functions. In this study, we investigated new lncRNAs in HIV-1-infected patients and studied the difference between the expression of lncRNAs associated with the time when ART treatment was started and controls.
Materials and methods
Data selection
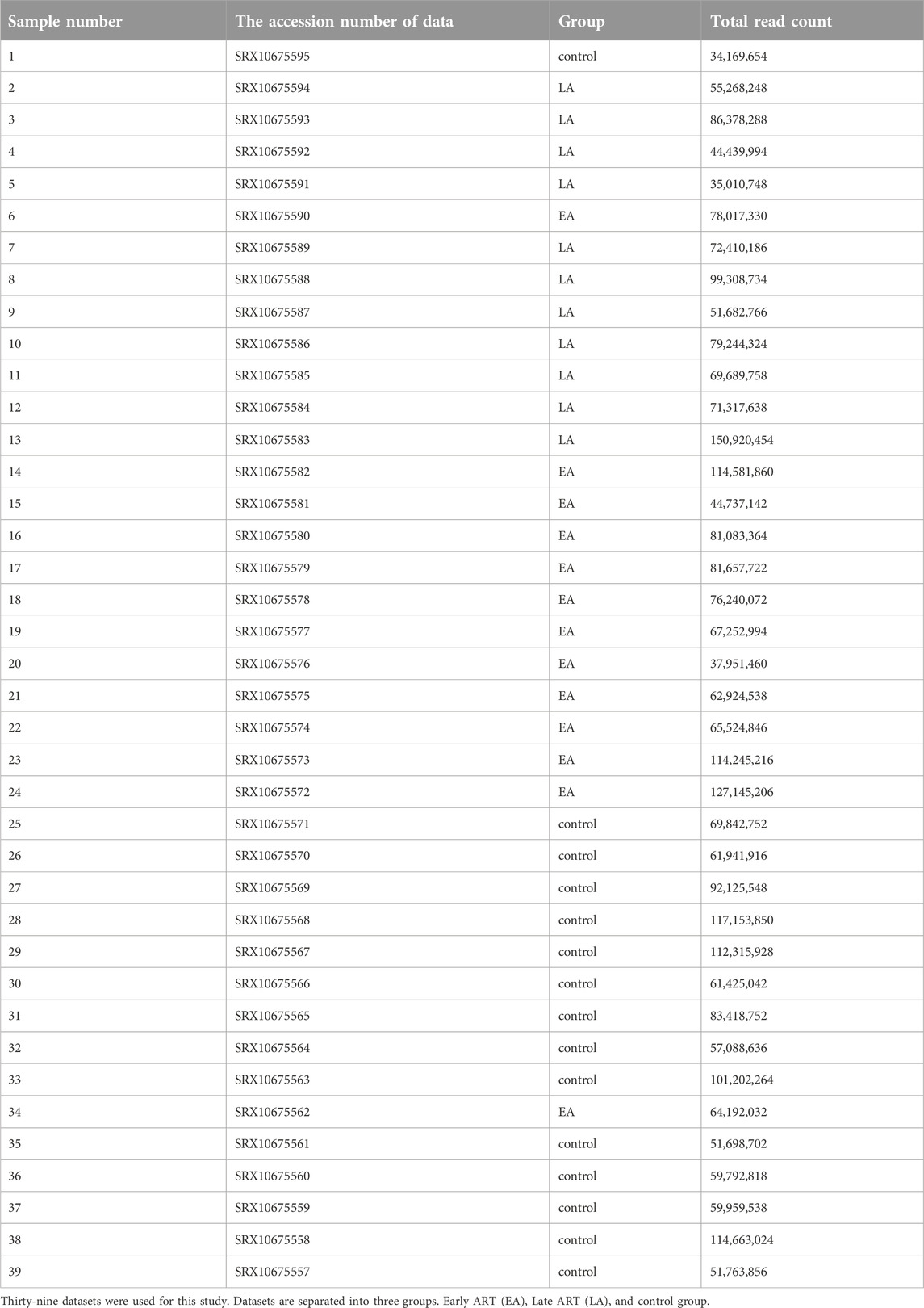
We investigated whole transcriptome data or total RNA-Seq studies. Sequence datasets were obtained from the GenBank data libraries1 under accession number PRJNA724934 (Petkov and Chiodi, 2021). Thirty-nine RNA-seq datasets in FASTQ format were downloaded using the dataset sample experiment.
lncRNA discovery
De novo assembly of datasets
Datasets were analyzed using CLC Genomics Workbench (v20) software. All 39 datasets underwent quality control and trimming. Additionally, reads less than 15 bp were automatically discarded through adapter trimming. The trimmed datasets were then de novo assembled, with a contig length threshold of 250 bp and a word size of 20.
Reference mapping
The human reference genome (GCF_000001405.40_GRCh38.p14_genomic) was obtained from NCBI2. The de novo assembly results were mapped against the annotated mRNA track of the reference genome using a length fraction of 0.8% and 95% identity. The unmapped reads were utilized for further analyses.
PFAM domain search
The unmapped contigs were translated into proteins using all six frames. Subsequently, the amino acid sequences underwent screening for PFAM domains, and any identified protein domain was subsequently excluded from the obtained data. The conserved protein domains were identified using the Pfam domain search algorithm used in the CLC Genomic Workbench.
ORF finding
The PFAM domain search results were uploaded to CPC3, and the encoding potential was determined. All contigs with coding potential were discarded, and the remaining contigs were used for further analyses.
BLASTx against protein databases
A complete protein database was created in the CLC Genomics Workbench (v20) using all the existing protein databases for humans and animals in NCBI and UniProt4. The BLASTx algorithm was used to check the similarity of the reads with known proteins (query genetic code: 1 standard, E-value <0.0001 and a maximum number of hit sequences = 20, word size: 3, matrix: BLOSUM62, gap cost: existence 11, extension 1, and a maximum number of hit sequences: 2). The complete animal protein RefSeq and PFAM databases were also used to find protein-coding reads. All the remaining contigs that were identified as proteins were sorted out. The remaining contigs were considered putative lncRNAs.
Annotation of putative lncRNAs
In order to distinguish novel lncRNAs from conserved lncRNAs, the FASTA format of the human lncRNA reference (Homo_sapiens.GRCh38) was downloaded from Ensembl5. Subsequently, the putative lncRNAs were annotated with the lncRNA reference sequence using the BLASTn program (p-value < 0.0001). Finally, a new lncRNA reference was created, consisting of both the conserved and novel lncRNAs. The lncRNA discovery method is summarized in Figure 1.
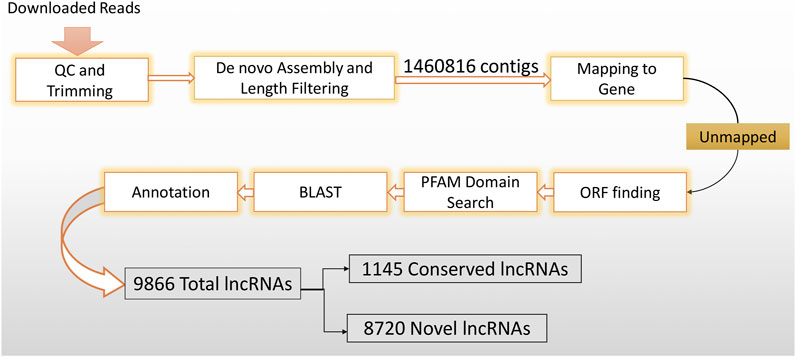
Figure 1. LncRNA discovery method flowchart. In order to get highly confident putative lncRNAs, downloaded datasets were imported into the CLC genomic workbench and subjected to rigorous filtering.
Expression and statistical analysis
In order to determine which lncRNAs were up- and downregulated in each sample, the previously generated trimmed datasets were mapped against the newly generated lncRNA reference (length fraction = 0.8, similarity fraction = 0.8, mismatch cost = 2, insertion cost = 3, deletion cost = 3, and the maximum number of hits for a read = 10. Expression values were determined as reads per kilobase of transcript per million reads mapped, RPKM) (Davati and Ghorbani, 2023). The mapped results for each sample were used for expression analysis. The expression analysis results were statistically analyzed using Empirical Analysis DGE. The analysis included the exact test comparison, and samples were compared to controls using FDR-corrected p-values. The data were then filtered to identify differentially expressed (DE) lncRNAs. LncRNAs with an EDGE test fold change (FC) of < -2 or >2 and an EDGE test FDR p-value correction <0.05 were maintained as DE lncRNAs. A heatmap was generated using heatmapper tools6 with default parameters (average linkage clustering methods and Euclidean distance measurement methods) to calculate Z-Score from the RPKMs of lncRNAs in samples.
LncRNA-miRNA interaction analysis
The mature miRNA sequences of Homo sapiens were extracted from the miRBase database7. Lnc-miRNA interactions were determined using psRNATarget8. Interaction networks for commonly up- and downregulated lncRNAs with mature miRNAs were then generated using Cytoscape (v3.9.1) software9.
Identification of the subcellular localization of lncRNAs
LncRNA sequences for the two common groups (upregulated and downregulated) were uploaded in FASTA format into LightGBM-LncLoc (LncRNA subcellular localization predictor based on LightGBM) (Lyu et al., 2023).
mRNA-lncRNA interaction and gene ontology analysis
The chromosome localization of upregulated and downregulated lncRNAs was determined using Geneious Prime (v.2019.1)10 for transcript annotation of ncRNA. The annotated sequences were further aligned with the human chromosome reference sequence utilizing the Blast annotation tools (with a maximum E-value of ≤0.0001). Subsequently, the results were converted to tracks using the track conversion tool. These track data were manually analyzed to identify genomic regions wherein genes were located adjacent to the lncRNAs, considering a distance of 10 kb. This manual analysis permitted a more detailed assessment of the relationships between lncRNAs and neighboring genes, leading to the gaining of multiple chromosomal regions.
Ensembl BioMart11 was important in transforming the numerous chromosomal regions acquired into matching gene IDs. This phase improved the accuracy and organized classification of the genes linked to the investigated lncRNAs.
The discovered genes were analyzed for enrichment in gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The STRING v.11.512 was used to do multiple protein searches in the human database, using the default parameters.
Results and discussion
Identification of putative lncRNAs in human CD4+ naïve T cells
A strong reference of human CD4+ naïve T-cell lncRNAs was generated using 39 datasets (Table 1) extracted from SRA. These datasets were previously collected for another study examining the effects of using ART in the early stages of infection on CD4+ naïve T cells. Overall, the RNA-seqs included 13 datasets from patients who received early ART (EA), 11 datasets from patients who received late ART (LA), and 15 datasets from healthy controls. To identify lncRNAs, we developed a highly restricted methodology. Briefly, 1,460,816 contigs were detected, consisting of coding and non-coding transcripts. After removing the reads related to the genes, coding potential, or protein domains, the remaining reads with protein-coding ability were also discarded. Finally, a total of 9,866 lncRNAs were discovered. Annotation distinguished 8,720 novel lncRNAs from 1,145 conserved lncRNAs. In a previous study, 17,235 lncRNAs were identified from peripheral blood mononuclear cells (PBMCs) from three EA and three healthy controls (Ma et al., 2021). The difference between the identified lncRNAs is likely due to the rigorous lncRNA discovery method we used and, more importantly, the fact that in our study, lncRNAs were identified only for naïve CD4+ T cells purified from PBMCs.
Differential expression of lncRNAs in EA and LA groups compared to control
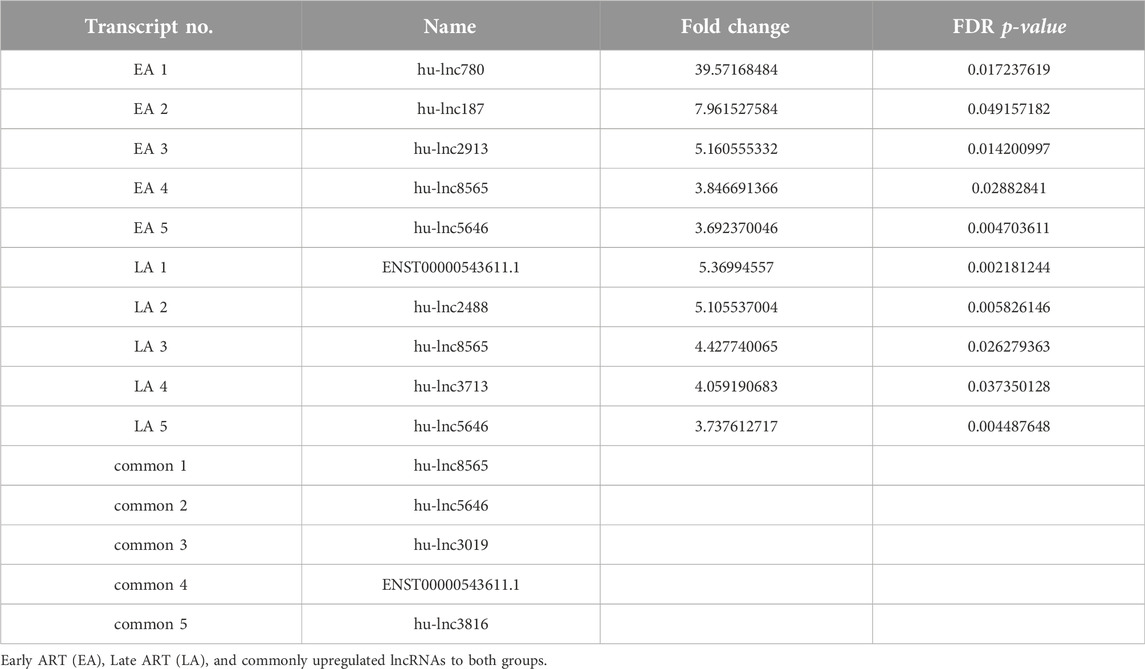
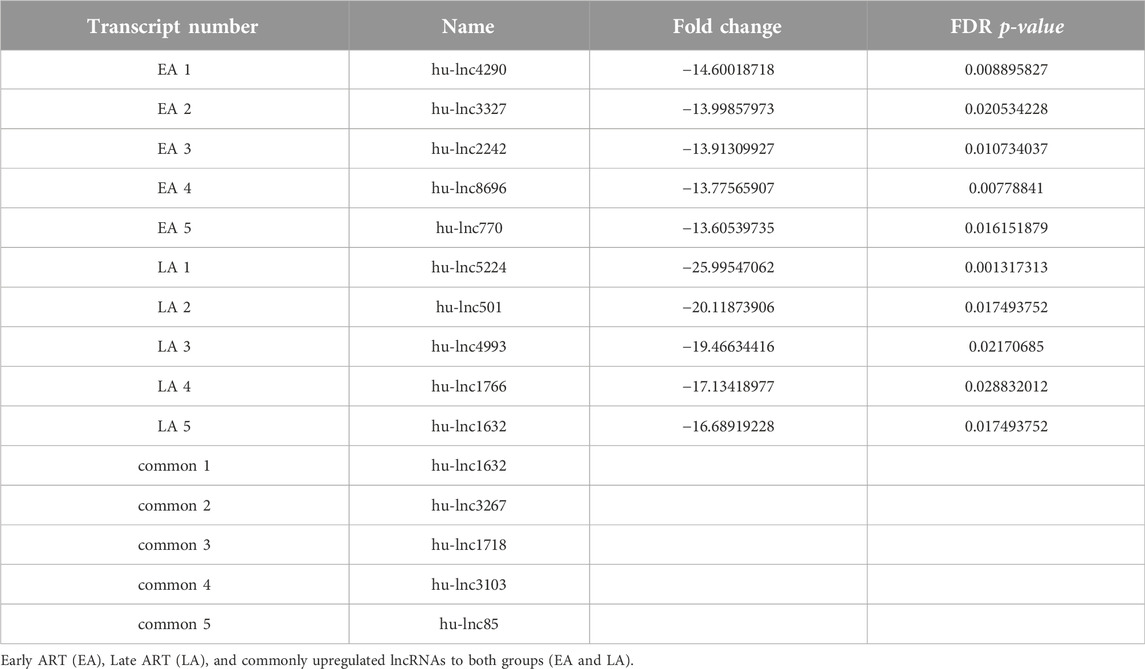
To gain a comprehensive understanding of differentially expressed (DE) lncRNAs in CD4+ T cells during treated HIV-1 infection, we compared 13 EA and 11 LA RefSeq transcriptomic datasets with 15 controls. Supplementary Data S1 includes an analysis of DE lncRNAs in both the EA and LA groups compared to the controls. After filtering, we confirmed that 20 lncRNAs were upregulated in the EA group, while 122 were downregulated compared to the control group. In the LA group, 23 lncRNAs were upregulated, and 151 were downregulated compared to the control group. We identified 20 downregulated and 9 upregulated lncRNAs that were common to both groups (EA and LA). Volcano plots for each group are shown in Figure 2 and a heat map for lncRNAs is provided in Figure 3. Ma et al. identified 36 upregulated and 206 downregulated lncRNAs in PBMCs from three ART-naive patients with early HIV-1 infection (within 180 days of HIV-1 infection) and three healthy controls (Ma et al., 2021). Trypsteen et al. detected 262 differentially expressed lncRNAs in cultured central memory T cells and bystander models compared to mock-infected cells. The majority of dysregulated lncRNAs were upregulated in both models (Trypsteen et al., 2019). Based on microarray analysis, Nair et al. identified 795 lncRNAs that were downregulated during active infection in U-937 monocytes infected with HIV-1 (in vitro) and 389 lncRNAs that were upregulated compared to uninfected controls (Nair et al., 2016). Our results are consistent with two of the mentioned studies regarding the expression pattern of DE lncRNAs. We also observed a higher number of downregulated lncRNAs compared to upregulated ones, which aligns with their findings. The difference in the number of DE lncRNAs observed in our study may be attributed to the influence of antiretroviral therapy (ART) on lncRNA expression, particularly in the LA group, where patients had undergone ART for an extended period. Furthermore, recent evidence suggests that the expression of lncRNAs is cell-type specific (Bjørklund et al., 2022). We restricted our study to naïve CD4+ T cells, which could be another reason for the differences in the number of differentially expressed (DE) lncRNAs. Additionally, some of the studies were performed in vitro, which could also influence the outcome. Two top-upregulated lncRNAs (hu-lnc5646 and hu-lnc8565) (Tables 2, 3) were common in both the EA and LA groups. Further investigation of these lncRNAs may provide us with biomarkers for HIV-1 infection. ENST00000543611.1 was the only conserved lncRNA among the top upregulated lncRNAs (Table 2). Among the identified lncRNAs, some belonged to already known lncRNAs such as ENTPD1 antisense RNA 1 (ENST00000665049.1), TRIM8 divergent transcript (ENST00000607967.1), and PRR34 antisense RNA 1 (ENST00000451166.5). More information on the DE lncRNAs is provided in Supplementary Data S1. One of the commonly upregulated lncRNAs is ENST00000661141.1, also known as long intergenic non-protein coding RNA 1278 (LINC01278). LINC01278 may play different roles in different tumors. One study indicated that LINC01278 contributes to the development of hepatocellular carcinoma (HCC) (Huang et al., 2020). Another study found that LINC01278, as a positive regulator of DNM3 expression, suppresses thyroid carcinoma progression (Lin et al., 2019). Although it is not clear how LINC01278 functions in HIV-1 positive patients, the upregulation of this lncRNA in both treatments seems to play a role in human response to ART. Hu-lnc780 in the EA group (Table 2) showed a significantly greater extent of upregulation, whereas hu-lnc5224 in the LA group (Table 3) exhibited a much greater extent of downregulation, making them attractive candidates for further investigation.
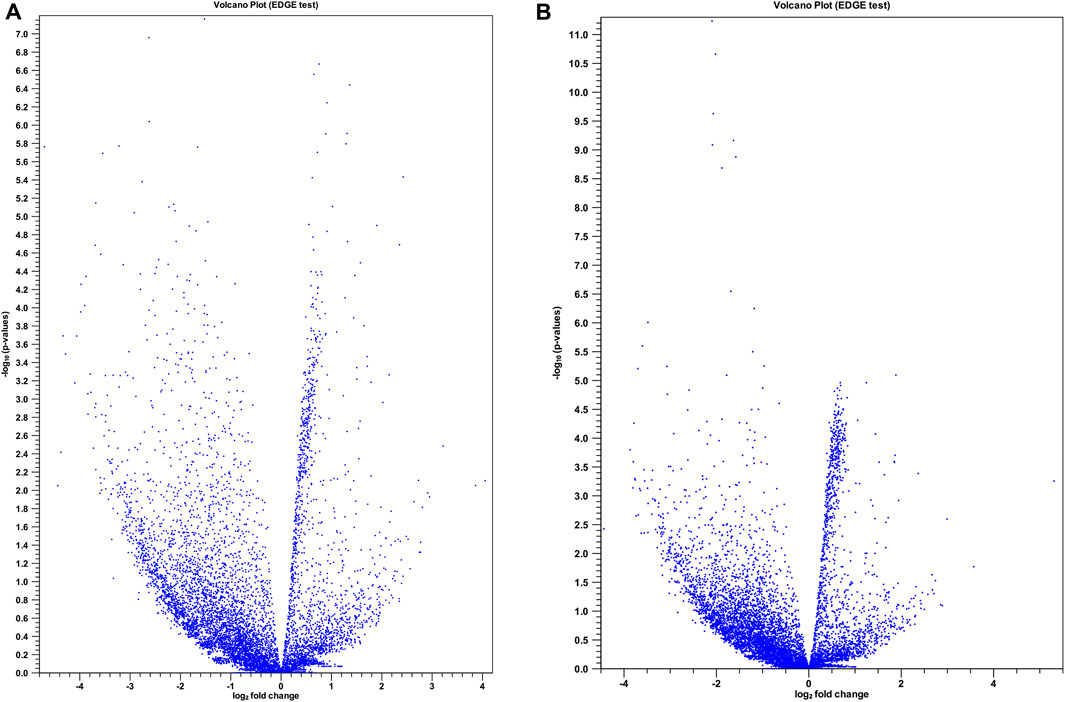
Figure 2. Volcano plots for differentially expressed (DE) lncRNAs in two treatment groups compared with control. (A) Control vs. late ART (LA) and (B) control vs. early ART (EA) with extraction and analysis of differential gene expression (EDGE) test. The x-axis shows the magnitude (log2 fold change) of DE lncRNAs and the y-axis shows significance (− log 10 adjusted p-value). Each blue spot represents a specific DE lncRNA.
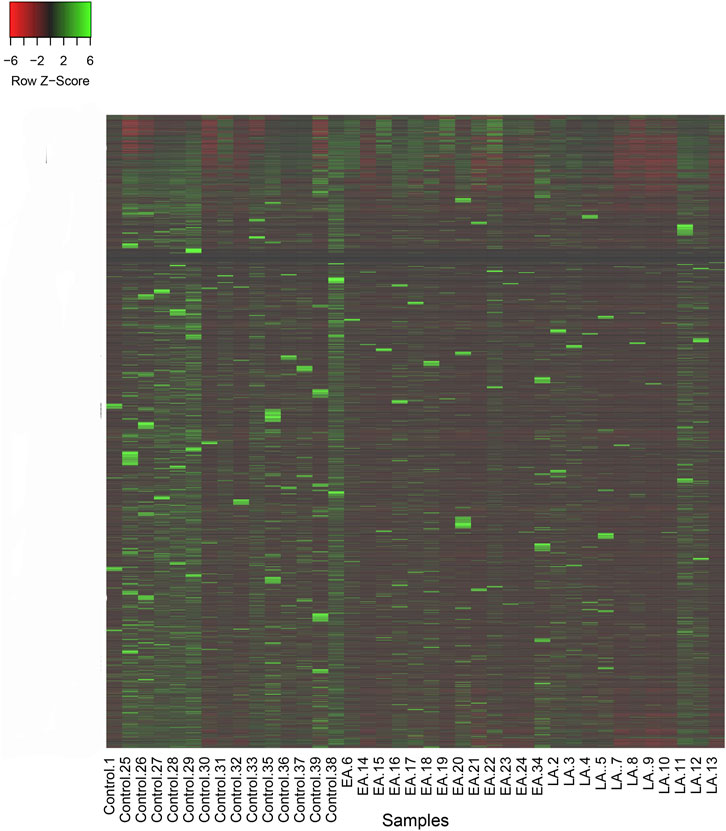
Figure 3. Heatmap of reads per kilobase of transcript per million reads mapped (RPKM) for lncRNAs in two treatment groups, late ART (LA) and early ART (EA), and control. Higher values of Z-Score for lncRNAs in each sample are shown in green and lower values are shown in red. Z-Score is calculated by: (RPKM in a sample of interest) - (Mean RPKM across all samples)/Standard Deviation.
LncRNAs affect miRNAs involved in tumor regulation
The extensive repertoire of miRNAs and their wide-ranging capacity to target various genes indicate the presence of an intricate regulatory network that modulates gene expression (Disner et al., 2021). Therefore, the mature miRNA sequences of H. sapiens were acquired from the miRBase database to anticipate these interactions. Then, the up-and downregulated lncRNAs for each treatment and the common up- and downregulated lncRNAs in both treatments were identified. Each group was submitted to psRNATarget8 for comparison with miRNA sequences from miRBase. Interaction networks of common up- and downregulated lncRNAs and mature miRNAs were then generated using Cytoscape software (v3.9.1). The top 50 interactions were selected to allow for more detailed visualization. Figure 4 shows the top 50 common lncRNA-miRNA interaction networks. A highly upregulated lncRNA (hu-lnc8565) was found to interact with at least nine different miRNAs (Figure 4A), including hsa-miR-4739, hsa-miR-6873-3p, hsa-miR-6838-3p, hsa-miR-4753-3p, hsa-miR-6501-5p, hsa-miR-6090, hsa-miR-6796-5p, hsa-miR-6085, and hsa-miR-6891-5p. ENST00000543611.1 was another highly upregulated lncRNA that showed interactions with several miRNAs (Figure 4A), such as hsa-miR-3911, hsa-miR-3147, hsa-miR-6640-5p, hsa-miR-4539, hsa-miR-6800-5p, and hsa-miR-6779-5p. A highly downregulated lncRNA (hu-lnc1632) also showed interactions with at least two miRNAs, including hsa-miR-6875-3p and hsa-miR-574-5p (Figure 4B). The results also showed that key miRNAs, including hsa-miR-574-5p, hsa-miR-765, hsa-miR-6165, and hsa-miR-1207-5p, can be controlled by commonly downregulated lncRNAs. Most of these miRNAs are thought to have targets involved in tumor prevention or development. Hsa-miR-574-5p has been shown to reduce liver metastasis in colon cancer by negatively regulating the expression of Metastasis-Associated in Colon Cancer 1, a biomarker of carcinogenesis and metastasis in more than 20 cancers (Cui et al., 2014; Wen et al., 2020). However, hsa-miR-574-5p was found to be a major factor associated with accelerated tumor growth in human lung cancer (Li et al., 2012). Hsa-miR-765, which is overexpressed in many human cancers, may act as a tumor suppressor and control arterial stiffness (Xie et al., 2016; Pallez et al., 2017; Zammit et al., 2018). It has been shown that hsa-miR-765 can increase cell growth by suppressing the expression of INPP4B in HCC (Liu et al., 2017). Hsa-miR-6165 is expressed in brain tumor-derived cell lines and primary brain tumor tissues. Its overexpression has been shown to downregulate the apoptosis-related genes PKD1 and DAGLA and induce NGFR, which functions as a tumor suppressor in most cancers (Parsi et al., 2012). Hsa-miR-1207-5p prevents gastric cancer development and invasion by targeting telomerase reverse transcriptase, ultimately leading to tumor senescence and death (Chen et al., 2014; Xu et al., 2015). It interacts with the 3′UTR of hTERT in gastric malignancies and is significantly downregulated in tissue samples from these tumors (Qin et al., 2015; Dinami et al., 2017). Moreover, hsa-miR-1207-5p was found to be strongly downregulated in breast cancer compared with healthy tissues (Dinami et al., 2017). Frequently upregulated lncRNAs also show interactions with important miRNAs such as hsa-miR-378i, hsa-miR-328-5p, hsa-miR-4753-3p, and hsa-miR-664b-5p. Hsa-miR-378i may act as a negative regulator in the progression of colon cancer by inducing apoptosis and preventing colon cancer cells from migrating and invading other tissues (Zeng et al., 2017).
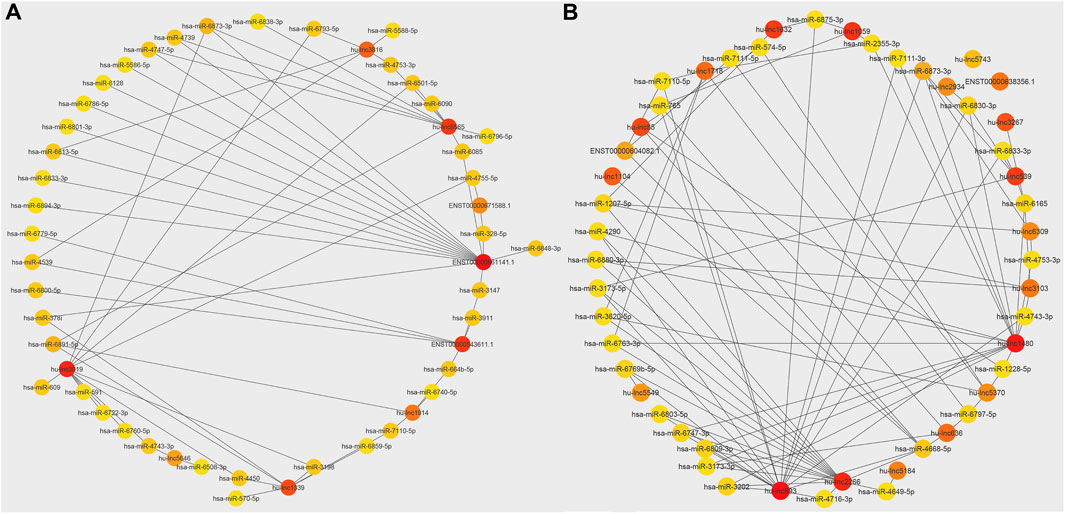
Figure 4. Network of interaction between top common lncRNAs and miRNAs. (A) Commonly upregulated lncRNAs and miRNAs interaction. (B) Commonly downregulated lncRNAs and miRNAs interaction. The colors of the circles, red, orange, and yellow, indicate the importance of interaction nodes ranging from high to low.
However, hsa-miR-378i has also been shown to increase cell survival, decrease caspase-3 activity, and promote tumorigenesis, angiogenesis, and neurogenesis. It can also induce neuroprotection by negatively regulating caspase-3-linked apoptosis (Meganathan et al., 2015; Zhang et al., 2016). The expression of hsa-miR-328-5p is closely associated with cell cycle progression, and numerous malignancies exhibit low expression of hsa-miR-328-5p (Wu et al., 2012). It has been established that hsa-miR-328-5p plays a role in the control of phagocytosis and bacterial survival, and its downregulation increases the clearance of pathogens in the lung. Downregulation of hsa-miR-4753-3p has been shown to target genes involved in immunologically significant signaling pathways, such as MAPK signaling, cytokine-cytokine receptor interactions, endocytosis, actin cytoskeleton regulation, focal adhesion, calcium signaling, chemokine signaling, and Wnt signaling (Budak et al., 2018). MiR-664 may function as a pro-viral miRNA that is essential for the virus to downregulate downstream target genes involved in viral defense systems in the host cell (Wolf et al., 2016). Given this information, it is clear that these miRNAs can alter the course of the disease, play an important role in the immune response, and affect cancer cells. The interaction between lncRNAs expressed in patients who have received long-term ART with these miRNAs suggests that ART can affect controlling cancer and regulating the immune system. The findings are important because ART therapeutics, such as protease inhibitors (PIs), have been proven to have potent antiangiogenic effects and cause regression of Kaposi sarcoma (Sgadari et al., 2002; Toschi et al., 2011). However, the exact role of lncRNAs in regulating miRNAs and how these interactions may affect cancer or the immune system remains unclear and requires further research.
Identification of the subcellular localization of lncRNAs
As with proteins, the function of lncRNAs depends on their subcellular localization (Chen, 2016). The subcellular microenvironment enables different functions of the same lncRNA by allowing interactions with different functional protein partners and action targets (Bridges et al., 2021). The results of our study showed that the most common DE lncRNAs were localized in the nucleus. Eight out of twenty common downregulated lncRNAs were localized in the nucleus, and twelve lncRNAs were located in the cytoplasm (Table 4). Furthermore, eight out of nine common upregulated lncRNAs were localized in the nucleus, and one lncRNA was localized in the ribosome (Table 5). In total, 16 lncRNAs were localized in the nucleus, 12 in the cytoplasm, and one lncRNA in the ribosome. Structurally, lncRNAs are preferentially localized in the nucleus, more tissue-specific, less highly expressed, and evolutionarily less conserved than mRNAs (Liu et al., 2022).
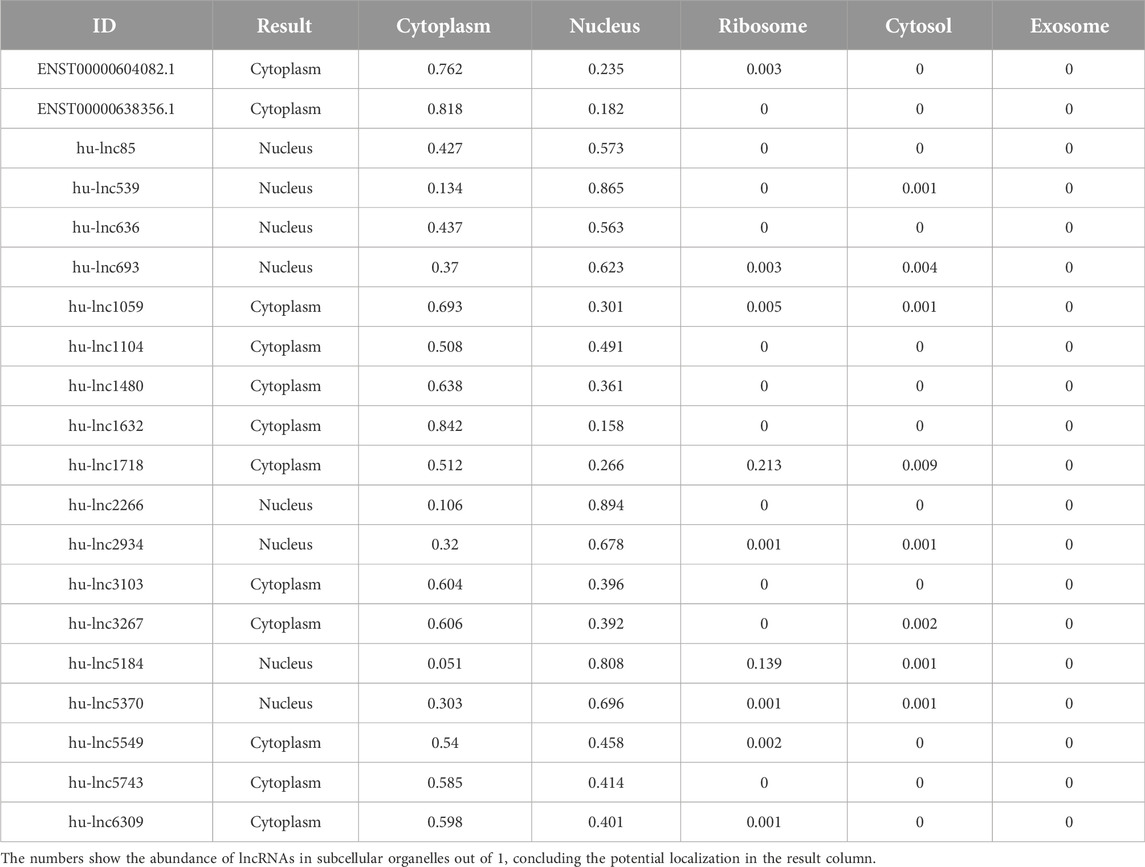
Table 4. Distribution of subcellular localization of commonly downregulated lncRNAs to both groups (EA and LA).
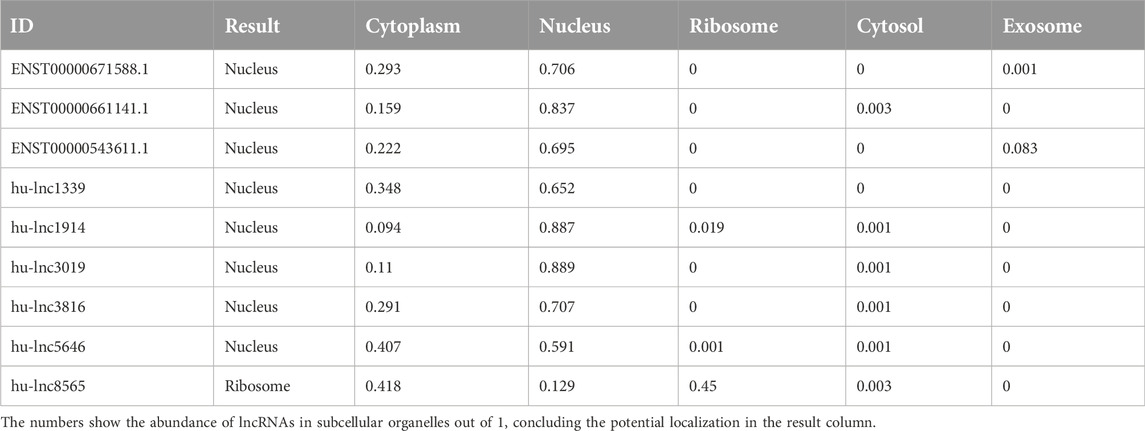
Table 5. Distribution of subcellular localization in commonly upregulated lncRNAs to both groups (EA and LA).
When localized in the nucleus, lncRNAs are involved in gene regulation at the epigenetic and transcriptional levels, including histone modifications, regulation of DNA methylation and chromatin remodeling, interaction with chromatin modification complexes, transcription factors, and proteins in the nucleus (Xing et al., 2021). Previous studies have shown that the substructural integrity of the nuclear paraspeckle, an important subcellular organelle for HIV-1 replication, is highly dependent on a lncRNA called NEAT1 (Zhang et al., 2013). The second group of lncRNAs must be exported to the cytoplasm to fulfill their regulatory roles (Chen, 2016). LncRNAs localized in the cytoplasm often function as scaffolds for protein complexes or as sponges for miRNAs acting at posttranscriptional and translational levels (Chen, 2016; Shen et al., 2020). The results may suggest that in HIV-1-positive patients receiving ART, both types of subcellular localization are expressed, affecting both nucleus mechanisms and cytoplasm mechanisms. The pathways by which lncRNAs function in the different subcellular localizations and whether these actions are beneficial for patient treatment are questions that remain unanswered.
GO enrichment analysis of the DE lncRNA-neighboring coding genes
We sought to investigate the potential targets of DE lncRNAs in cis-regulatory elements to shed light on their putative roles. Our analysis focused on identifying target genes of DE lncRNAs in CD4+ T cells from HIV-1 infected + ART-treated patients compared to controls. We acknowledge that our research included a total of 20 downregulated lncRNAs and 9 upregulated lncRNAs out of 9,866 lncRNAs which were down and upregulated as common in both treatments (Supplementary Data S1). We identified 4,209 transcripts in DE lncRNA-neighboring.
Subsequently, Gene Ontology (GO) analysis was performed to explore the functional implications of transcripts in DE lncRNA-neighboring. Comprehensive details on the terms enriched in the GO are available in Supplementary Data S2.
In our analysis, we identified enrichment (p-value < 0.05), in twelve Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Table 6). Notably, three major KEGG pathways were significantly enriched, namely, axon guidance (p-value = 0.0139), PI3K-Akt signaling pathway (p-value = 0.0398), and MAPK signaling pathway (p-value = 0.0066). Previous studies have linked impairment of host axon guidance and these signaling pathways to neurological manifestations in HIV-1 patients (Zhou et al., 2012; Scutari et al., 2017; Ojeda-Juárez and Kaul, 2021). In addition, our analysis identified terms linked to signaling pathways related to neurodegenerative diseases, including the MAPK signaling pathway, axon guidance pathway, and phosphorylative mechanism/signaling pathway. This supports the concept that these pathways play a noteworthy role in the development of HIV-1-associated dementia (HAD) (Zhou et al., 2012). Furthermore, it is important to note that earlier research has shown that some initial HIV-1 protease inhibitors might hinder the activation of Akt, indicating a possible connection to the pathways we discovered in our investigation (Plastaras et al., 2008). However, it is essential to use caution when interpreting this outcome, and more research is necessary to establish a definitive association and understand the broader implications of these findings.
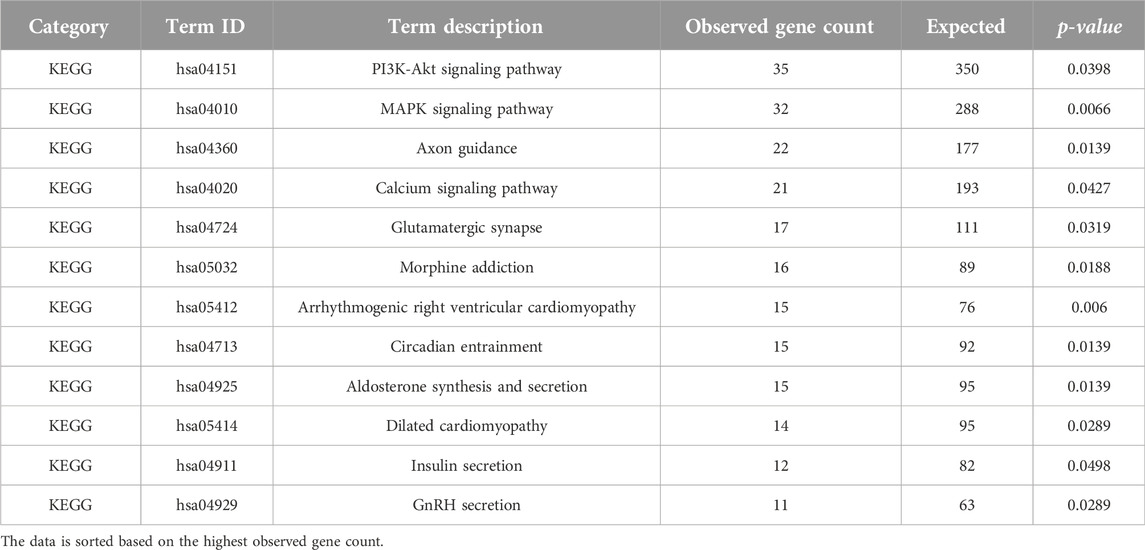
Table 6. KEGG pathways analysis (p-value < 0.05) of the determined DE lncRNA neighboring genes using STRING ver. 11.512.
Notably, our study revealed a significant enrichment of the glutamatergic synapse pathway (p-value = 0.0319). This observation suggests a potential association between CD4+ T cells and brain synaptic plasticity, which may have broader implications for cognitive functions (Zarif et al., 2018). It is crucial to accept that while our findings indicate a potential connection, more investigation is necessary to prove a cause-and-effect relationship. Prior research has shown that HIV-1 and viral proteins can interfere with the transmission of glutamate at synapses, which might lead to cognitive impairment (Ru and Tang, 2017; Gorska and Eugenin, 2020). Additionally, recent research has hinted at potential additive negative effects on the glutamatergic system when ART is combined with HIV-1 infection, further suggesting a connection to neuronal signaling (Gorska and Eugenin, 2020). These connections need thoughtful examination and deserve more experimental verification to completely understand their importance. While our analysis encompasses various pathways, we acknowledge the central focus of our study on CD4+ T cells and their role in the context of HIV-1 infection and ART treatment. We recognize the relevance of other studies that investigate the interaction of HIV-1 infection, ART, and lncRNAs in cells of the central nervous system, but for this chapter, we will concentrate on CD4+ T cells and their potential effects on relevant biological processes in these cells.
Neurotoxicity in HIV-1-infected patients is believed to be mediated by HIV-1 proteins, such as gp120 and transactivator of transcription (Tat), as well as other products released by infected cells (Fitting et al., 2008). Our analysis of biological functions (BF) (Table 7) revealed enrichment in terms related to protein binding (p-value = 0.009), enzyme binding (p-value = 0.038), enzyme regulatory activity (p-value = 0.0191), and cytoskeletal protein binding (p-value = 0.0133). The role of cytoskeletal proteins, such as actin and profilin 1, has been implicated in the efficient cell-to-cell spread of HIV-1 (Kadiu et al., 2007; Taylor et al., 2011). Additionally, ion channel activity, including potassium channel activity and calcium channel activity, emerged as enriched BF terms. Previous studies have demonstrated the significance of ion channels in HIV-1 replication and gene expression (Choi et al., 1998; Ohno, 2018; Dubey et al., 2019).
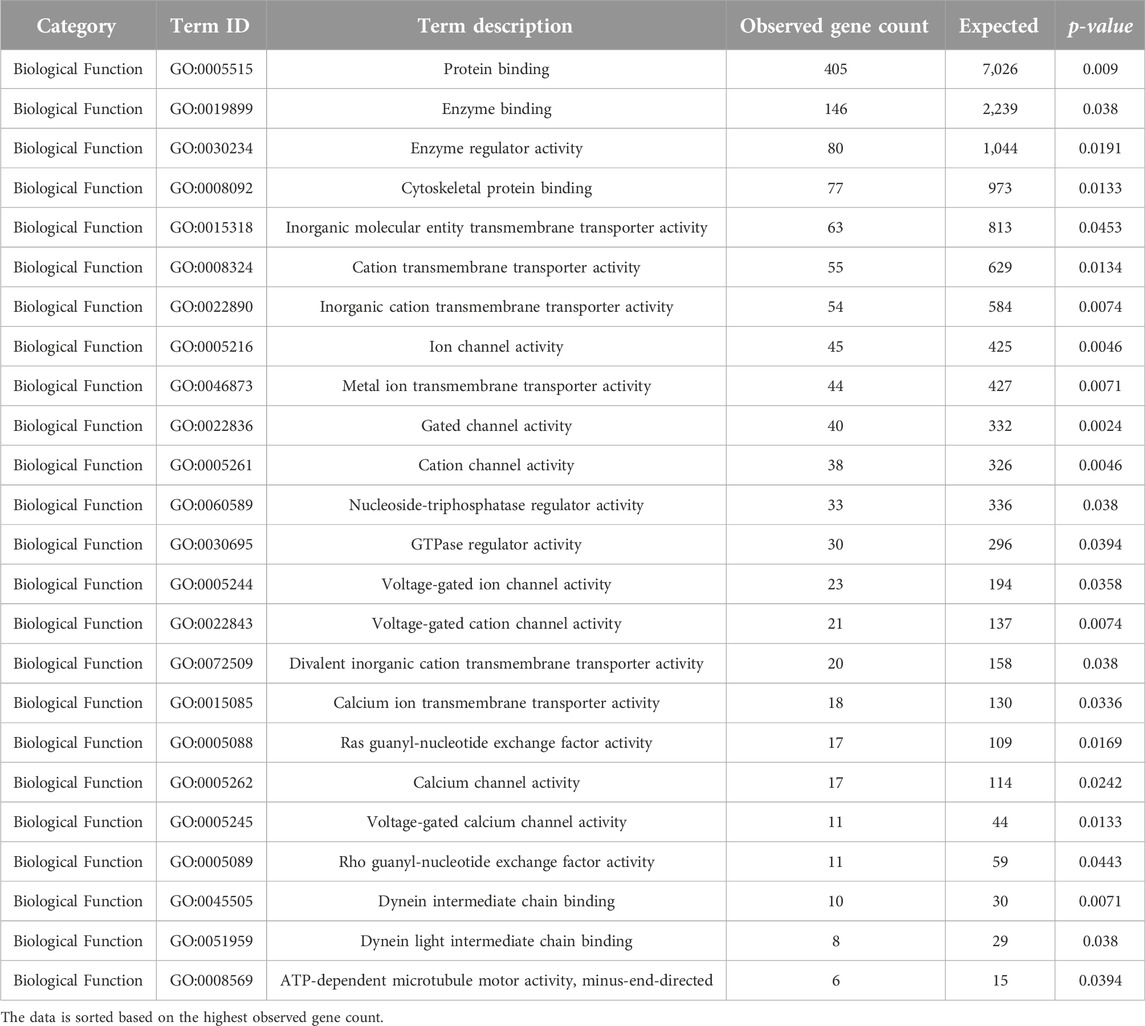
Table 7. GO analysis (Biological Function) (p-value < 0.05) of the determined DE lncRNA neighboring genes using STRING ver. 11.512.
Moreover, our analysis suggested a potential link between HIV-1 infection and early anatomical and functional changes in relevant brain regions, with implications for neuronal connectivity and glutamate release (Hu, 2016). We also highlight a study indicating that Tat-mediated upregulation of a specific long non-coding RNA (lncRNA), LOC102549805 (lncRNA-U1), disrupts neuronal homeostasis, further supporting the notion that lncRNAs may play a role in regulating ion channels and cellular functions in the context of HIV-1 infection (Torkzaban et al., 2020).
In summary, our analysis, while encompassing various pathways and potential targets of lncRNAs, is centered on the context of CD4+ T cells, HIV-1 infection, and ART treatment. Our findings suggest a complex interplay between lncRNAs, biological processes, and pathways that warrant further investigation to fully comprehend their mechanistic underpinnings.
Conclusion
In this study, we showed a thorough investigation into the role of lncRNAs in individuals infected with HIV-1 at various stages of infection and in response to ART. Our results produced a substantial set of 9,866 lncRNAs, separating 8,720 novel lncRNAs from 1,145 conserved counterparts. Particularly, we focused on the specific context of naïve CD4+ T cells purified from PBMCs, contributing to a more defined characterization. Comparison with previous investigations uncovered variations in identified lncRNAs, attributable to thorough discovery methodologies and the special focus on naïve CD4+ T cells. We detected characteristic expression patterns in patients receiving early and late ART, with a higher prevalence of downregulated lncRNAs. This aligns with earlier observations and could be attributed to the effect of prolonged ART and the cell-type specificity of lncRNA expression. Besides, two prominent lncRNAs, hu-lnc5646 and hu-lnc8565, were consistently upregulated in both early and late ART groups, warranting additional analysis as potential biomarkers for HIV-1 infection. Furthermore, the complicated interaction networks between identified lncRNAs and specific miRNAs underscored their regulatory potential, specifically in the context of ART. Our subcellular localization analysis emphasized a significant proportion of lncRNAs in the nucleus, proposing their involvement in epigenetic and transcriptional regulation. Notably, the differential localization across treatments implies complicated functions manipulated by the subcellular microenvironment. The cis-regulatory analysis uncovered potential targets of these lncRNAs, indicating their known roles in crucial biological functions.
The identification of pathways associated with axon guidance, PI3K-Akt signaling, and MAPK signaling in our work suggests possible links to neurological symptoms in HIV-1 patients. The results of our study provide insight into the potential role of lncRNAs in regulating ion channel activity, which might have significant implications for cellular function. The recognition of KEGG pathways, such as the glutamatergic synapse, highlights the crucial involvement of CD4+ T cells in synaptic plasticity, which has significance for HIV-1-related neurocognitive diseases. Significantly, our research offers a thorough summary of the regulatory network that involves lncRNAs in the context of HIV-1 infection and antiretroviral therapy (ART). Although we have seen connections between identified lncRNAs and certain pathways, it is important to use caution since our work does not show direct causality or interactions. The identified lncRNAs, their interactions with miRNAs, and their localization offer constructive insights into their possible roles in modulating immune responses and cancer-related pathways. Future research endeavors should plan to reveal the detailed mechanisms by which lncRNAs function in different subcellular compartments and their effect on disease progression and treatment outcomes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
NF and AG design and analysis of data. TK and NM-D wrote the draft. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Next-Generation EU - Italian NRRP, Mission 4, Component 2, Investment 1.5, call for the creation and strengthening of ‘Innovation Ecosystems’, building ‘Territorial R&D Leaders’ (Directorial Decree n. 2021/3277) - project Tech4You - Technologies for climate change adaptation and quality of life improvement, n. ECS0000009. Also, this work is based on research funded by Iran National Science Foundation (INSF) under project No. 4015898.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author disclaimer
This work reflects only the authors’ views and opinions, neither the Ministry for University and Research nor the European Commission can be considered responsible for them.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2024.11572/full#supplementary-material
Footnotes
1https://www.ncbi.nlm.nih.gov/genbank/
5https://useast.ensembl.org/index.html
6http://www.heatmapper.ca/expression/
11http://asia.ensembl.org/biomart/martview
References
Bale, M. J., and Kearney, M. F. (2019). Review: HIV-1 phylogeny during suppressive antiretroviral therapy. Curr. Opin. HIV AIDS 14 (3), 188–193. doi:10.1097/coh.0000000000000535
Bandera, A., Gori, A., Clerici, M., and Sironi, M. (2019). Phylogenies in ART: HIV reservoirs, HIV latency and drug resistance. Curr. Opin. Pharmacol. 48, 24–32. doi:10.1016/j.coph.2019.03.003
Bjørklund, S. S., Aure, M. R., Häkkinen, J., Vallon-Christersson, J., Kumar, S., Evensen, K. B., et al. (2022). Subtype and cell type specific expression of lncRNAs provide insight into breast cancer. Commun. Biol. 5 (1), 834. doi:10.1038/s42003-022-03559-7
Boliar, S., Gludish, D. W., Jambo, K. C., Kamng'ona, R., Mvaya, L., Mwandumba, H. C., et al. (2019). Inhibition of the lncRNA SAF drives activation of apoptotic effector caspases in HIV-1-infected human macrophages. Proc. Natl. Acad. Sci. U. S. A. 116 (15), 7431–7438. doi:10.1073/pnas.1818662116
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA localization and function. J. Cell Biol. 220 (2), e202009045. doi:10.1083/jcb.202009045
Budak, F., Bal, S. H., Tezcan, G., Akalın, E. H., Yılmaz, A., Hız, P., et al. (2018). The microRNA expression signature of CD4+ T cells in the transition of brucellosis into chronicity. PLoS One 13 (6), e0198659. doi:10.1371/journal.pone.0198659
Campos Coelho, A. V., Moura, R. R. D., and Crovella, S. (2020). Reanalysis of gene expression profiles of CD4+ T cells treated with HIV-1 latency reversal agents. Microorganisms. 8 (10), 1505. doi:10.3390/microorganisms8101505
Chao, T.-C., Zhang, Q., Li, Z., Tiwari, S. K., Qin, Y., Yau, E., et al. (2019). The long noncoding RNA HEAL regulates HIV-1 replication through epigenetic regulation of the HIV-1 promoter. mBio 10 (5), e02016–e02019. doi:10.1128/mbio.02016-19
Chen, H., and Shan, G. (2020). The physiological function of long-noncoding RNAs. Non-coding RNA Res. 5 (4), 178–184. doi:10.1016/j.ncrna.2020.09.003
Chen, L., Lü, M. H., Zhang, D., Hao, N. B., Fan, Y. H., Wu, Y. Y., et al. (2014). miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 5 (1), e1034. doi:10.1038/cddis.2013.553
Chen, L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41 (9), 761–772. doi:10.1016/j.tibs.2016.07.003
Choi, B., Gatti, P. J., Haislip, A. M., Fermin, C. D., and Garry, R. F. (1998). Role of potassium in human immunodeficiency virus production and cytopathic effects. Virology 247 (2), 189–199. doi:10.1006/viro.1998.9251
Chopra, S., Kakar, A., and Gogia, A. (2019). Advanced HIV: diagnosis, treatment, and prevention. Curr. Med. Res. Pract. 9 (5), 201–202. doi:10.1016/j.cmrp.2019.09.003
Cui, Z., Tang, J., Chen, J., and Wang, Z. (2014). Hsa-miR-574-5p negatively regulates MACC-1 expression to suppress colorectal cancer liver metastasis. Cancer Cell Int. 14 (1), 47. doi:10.1186/1475-2867-14-47
Davati, N., and Ghorbani, A. (2023). Discovery of long non-coding RNAs in Aspergillus flavus response to water activity, CO2 concentration, and temperature changes. Sci. Rep. 13 (1), 10330. doi:10.1038/s41598-023-37236-4
De Boer, R. J., Homann, D., and Perelson, A. S. (2003). Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J. Immunol. 171 (8), 3928–3935. doi:10.4049/jimmunol.171.8.3928
Dinami, R., Buemi, V., Sestito, R., Zappone, A., Ciani, Y., Mano, M., et al. (2017). Epigenetic silencing of miR-296 and miR-512 ensures hTERT dependent apoptosis protection and telomere maintenance in basal-type breast cancer cells. Oncotarget 8 (56), 95674–95691. doi:10.18632/oncotarget.21180
Disner, G. R., Falcão, M. A. P., Lima, C., and Lopes-Ferreira, M. (2021). In silico target prediction of overexpressed microRNAs from LPS-challenged zebrafish (Danio rerio) treated with the novel anti-inflammatory peptide TnP. Int. J. Mol. Sci. 22 (13), 7117. doi:10.3390/ijms22137117
Dubey, R. C., Mishra, N., and Gaur, R. (2019). G protein-coupled and ATP-sensitive inwardly rectifying potassium ion channels are essential for HIV entry. Sci. Rep. 9 (1), 4113–4119. doi:10.1038/s41598-019-40968-x
Fitting, S., Booze, R. M., Hasselrot, U., and Mactutus, C. F. (2008). Differential long-term neurotoxicity of HIV-1 proteins in the rat hippocampal formation: A design-based stereological study. Hippocampus 18 (2), 135–147. doi:10.1002/hipo.20376
Gorska, A. M., and Eugenin, E. A. (2020). The glutamate system as a crucial regulator of CNS toxicity and survival of HIV reservoirs. Front. Cell. Infect. Microbiol. 10, 261. doi:10.3389/fcimb.2020.00261
Hu, X. T. (2016). HIV-1 tat-mediated calcium dysregulation and neuronal dysfunction in vulnerable brain regions. Curr. Drug Targets 17 (1), 4–14. doi:10.2174/1389450116666150531162212
Huang, W.-J., Tian, X.-P., Bi, S.-X., Zhang, S.-R., He, T.-S., Song, L.-Y., et al. (2020). The β-catenin/TCF-4-LINC01278-miR-1258-Smad2/3 axis promotes hepatocellular carcinoma metastasis. Oncogene 39 (23), 4538–4550. doi:10.1038/s41388-020-1307-3
Kadiu, I., Ricardo-Dukelow, M., Ciborowski, P., and Gendelman, H. E. (2007). Cytoskeletal protein transformation in HIV-1-Infected macrophage giant cells. J. Immunol. 178 (10), 6404–6415. doi:10.4049/jimmunol.178.10.6404
Leyre, L., Kroon, E., Vandergeeten, C., Sacdalan, C., Colby, D. J., Buranapraditkun, S., et al. (2020). Abundant HIV-infected cells in blood and tissues are rapidly cleared upon ART initiation during acute HIV infection. Sci. Transl. Med. 12 (533), eaav3491. doi:10.1126/scitranslmed.aav3491
Li, Q., Li, X., Guo, Z., Xu, F., Xia, J., Liu, Z., et al. (2012). MicroRNA-574-5p was pivotal for TLR9 signaling enhanced tumor progression via down-regulating checkpoint suppressor 1 in human lung cancer. PLoS One 7 (11), e48278. doi:10.1371/journal.pone.0048278
Lin, S., Tan, L., Luo, D., Peng, X., Zhu, Y., and Li, H. (2019). Linc01278 inhibits the development of papillary thyroid carcinoma by regulating miR-376c-3p/DNM3 axis. Cancer Manag. Res. 11, 8557–8569. doi:10.2147/cmar.s217886
Liu, L., Cai, X., Liu, E., Tian, X., and Tian, C. (2017). MicroRNA-18a promotes proliferation and metastasis in hepatocellular carcinoma via targeting KLF4. Oncotarget 8 (40), 68263–68269. doi:10.18632/oncotarget.19293
Liu, X., Xu, M., Li, P., Zhang, W., Zeng, L. H., Yang, Y., et al. (2022). Roles of lncRNAs in the transcription regulation of HIV-1. Biomed. J. 45 (4), 580–593. doi:10.1016/j.bj.2022.03.012
Lyu, J., Zheng, P., Qi, Y., and Huang, G. (2023). LightGBM-LncLoc: a LightGBM-based computational predictor for recognizing long non-coding RNA subcellular localization. Mathematics 11 (3), 602. doi:10.3390/math11030602
Ma, L., Zhang, H., Zhang, Y., Li, H., An, M., Zhao, B., et al. (2021). Integrated analysis of lncRNA, miRNA and mRNA profiles reveals potential lncRNA functions during early HIV infection. J. Transl. Med. 19 (1), 135. doi:10.1186/s12967-021-02802-9
Margolis, D. M., Archin, N. M., Cohen, M. S., Eron, J. J., Ferrari, G., Garcia, J. V., et al. (2020). Curing HIV: seeking to target and clear persistent infection. Cell 181 (1), 189–206. doi:10.1016/j.cell.2020.03.005
Meganathan, K., Jagtap, S., Srinivasan, S. P., Wagh, V., Hescheler, J., Hengstler, J., et al. (2015). Neuronal developmental gene and miRNA signatures induced by histone deacetylase inhibitors in human embryonic stem cells. Cell Death Dis. 6 (5), e1756. doi:10.1038/cddis.2015.121
Moran, V. A., Perera, R. J., and Khalil, A. M. (2012). Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 40 (14), 6391–6400. doi:10.1093/nar/gks296
Morcilla, V., Bacchus-Souffan, C., Fisher, K., Horsburgh, B. A., Hiener, B., Wang, X. Q., et al. (2021). HIV-1 genomes are enriched in memory CD4+ T-cells with short half-lives. mBio 12 (5), e0244721–21. doi:10.1128/mbio.02447-21
Nair, M., Sagar, V., and Pilakka-Kanthikeel, S. (2016). Gene-expression reversal of lncRNAs and associated mRNAs expression in active vs latent HIV infection. Sci. Rep. 6 (1), 34862. doi:10.1038/srep34862
Ohno, Y. (2018). Astrocytic Kir4.1 potassium channels as a novel therapeutic target for epilepsy and mood disorders. Neural Regen. Res. 13 (4), 651–652. doi:10.4103/1673-5374.230355
Ojeda-Juárez, D., and Kaul, M. (2021). Transcriptomic and genetic profiling of HIV-associated neurocognitive disorders. Front. Mol. Biosci. 8, 721954. doi:10.3389/fmolb.2021.721954
Ørom, U. A., Derrien, T., Beringer, M., Gumireddy, K., Gardini, A., Bussotti, G., et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143 (1), 46–58. doi:10.1016/j.cell.2010.09.001
Pallez, D., Gardès, J., and Pasquier, C. (2017). Prediction of miRNA-disease associations using an evolutionary tuned latent semantic analysis. Sci. Rep. 7 (1), 10548. doi:10.1038/s41598-017-10065-y
Parsi, S., Soltani, B. M., Hosseini, E., Tousi, S. E., and Mowla, S. J. (2012). Experimental verification of a predicted intronic microRNA in human NGFR gene with a potential pro-apoptotic function. PLoS One 7 (4), e35561. doi:10.1371/journal.pone.0035561
Peng, X., Gralinski, L., Armour, C. D., Ferris, M. T., Thomas, M. J., Proll, S., et al. (2010). Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 1 (5), e00206–e00210. doi:10.1128/mbio.00206-10
Petkov, S., and Chiodi, F. (2021). Distinct transcriptomic profiles of naïve CD4+ T cells distinguish HIV-1 infected patients initiating antiretroviral therapy at acute or chronic phase of infection. Genomics. 113 (6), 3487–3500. doi:10.1016/j.ygeno.2021.08.014
Petkov, S., and Chiodi, F. (2022). Impaired CD4+ T cell differentiation in HIV-1 infected patients receiving early anti-retroviral therapy. Genomics 114 (3), 110367. doi:10.1016/j.ygeno.2022.110367
Plastaras, J. P., Vapiwala, N., Ahmed, M. S., Gudonis, D., Cerniglia, G. J., Feldman, M. D., et al. (2008). Validation and toxicity of PI3K/Akt pathway inhibition by HIV protease inhibitors in humans. Cancer Biol. Ther. 7 (5), 628–635. doi:10.4161/cbt.7.5.5728
Qin, Y. Z., Xie, X. C., Liu, H. Z., Lai, H., Qiu, H., and Ge, L. Y. (2015). Screening and preliminary validation of miRNAs with the regulation of hTERT in colorectal cancer. Oncol. Rep. 33 (6), 2728–2736. doi:10.3892/or.2015.3892
Ru, W., and Tang, S.-J. (2017). HIV-associated synaptic degeneration. Mol. Brain 10 (1), 40. doi:10.1186/s13041-017-0321-z
Scutari, R., Alteri, C., Perno, C. F., Svicher, V., and Aquaro, S. (2017). The role of HIV infection in neurologic injury. Brain Sci. 7 (4), 38. doi:10.3390/brainsci7040038
Sgadari, C., Barillari, G., Toschi, E., Carlei, D., Bacigalupo, I., Baccarini, S., et al. (2002). HIV protease inhibitors are potent anti-angiogenic molecules and promote regression of Kaposi sarcoma. Nat. Med. 8 (3), 225–232. doi:10.1038/nm0302-225
Shen, L., Wu, C., Zhang, J., Xu, H., Liu, X., Wu, X., et al. (2020). Roles and potential applications of lncRNAs in HIV infection. Int. J. Infect. Dis. 92, 97–104. doi:10.1016/j.ijid.2020.01.006
Taylor, M. P., Koyuncu, O. O., and Enquist, L. W. (2011). Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 9 (6), 427–439. doi:10.1038/nrmicro2574
Torkzaban, B., Natarajaseenivasan, K., Mohseni Ahooyi, T., Shekarabi, M., Amini, S., Langford, T. D., et al. (2020). The lncRNA LOC102549805 (U1) modulates neurotoxicity of HIV-1 Tat protein. Cell Death Dis. 11 (10), 835. doi:10.1038/s41419-020-03033-4
Toschi, E., Sgadari, C., Malavasi, L., Bacigalupo, I., Chiozzini, C., Carlei, D., et al. (2011). Human immunodeficiency virus protease inhibitors reduce the growth of human tumors via a proteasome-independent block of angiogenesis and matrix metalloproteinases. Int. J. Cancer 128 (1), 82–93. doi:10.1002/ijc.25550
Trypsteen, W., White, C. H., Mukim, A., Spina, C. A., De Spiegelaere, W., Lefever, S., et al. (2019). Long non-coding RNAs and latent HIV - a search for novel targets for latency reversal. PLoS One 14 (11), e0224879. doi:10.1371/journal.pone.0224879
Wang, J., Zhang, Y., Li, Q., Zhao, J., Yi, D., Ding, J., et al. (2019). Influenza virus exploits an interferon-independent lncRNA to preserve viral RNA synthesis through stabilizing viral RNA polymerase PB1. Cell Rep. 27 (11), 3295–3304.e4. doi:10.1016/j.celrep.2019.05.036
Wen, J., Xie, Y., Zhang, Y., Li, J., Li, J., Zhang, Y., et al. (2020). MACC1 contributes to the development of osteosarcoma through regulation of the HGF/c-Met pathway and microtubule stability. Front. Cell Dev. Biol. 8, 825. doi:10.3389/fcell.2020.00825
Wolf, S., Wu, W., Jones, C., Perwitasari, O., Mahalingam, S., and Tripp, R. A. (2016). MicroRNA regulation of human genes essential for influenza A (H7N9) replication. PLoS One 11 (5), e0155104. doi:10.1371/journal.pone.0155104
Wu, Z., Sun, L., Wang, H., Yao, J., Jiang, C., Xu, W., et al. (2012). MiR-328 expression is decreased in high-grade gliomas and is associated with worse survival in primary glioblastoma. PLoS One 7 (10), e47270. doi:10.1371/journal.pone.0047270
Xie, B. H., He, X., Hua, R. X., Zhang, B., Tan, G. S., Xiong, S. Q., et al. (2016). Mir-765 promotes cell proliferation by downregulating INPP4B expression in human hepatocellular carcinoma. Cancer Biomarkers 16 (3), 405–413. doi:10.3233/cbm-160579
Xing, C., Sun, S. G., Yue, Z. Q., and Bai, F. (2021). Role of lncRNA LUCAT1 in cancer. Biomed. Pharmacother. 134, 111158. doi:10.1016/j.biopha.2020.111158
Xu, X., Chen, W., Miao, R., Zhou, Y., Wang, Z., Zhang, L., et al. (2015). miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget 6 (6), 3988–4004. doi:10.18632/oncotarget.2905
Zammit, V., Baron, B., and Ayers, D. (2018). MiRNA influences in neuroblast modulation: an introspective analysis. Genes. (Basel) 9 (1), 26. doi:10.3390/genes9010026
Zarif, H., Hosseiny, S., Paquet, A., Lebrigand, K., Arguel, M. J., Cazareth, J., et al. (2018). CD4(+) T cells have a permissive effect on enriched environment-induced Hippocampus synaptic plasticity. Front. Synaptic Neurosci. 10, 14. doi:10.3389/fnsyn.2018.00014
Zeng, M., Zhu, L., Li, L., and Kang, C. (2017). miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell. Mol. Biol. Lett. 22, 12. doi:10.1186/s11658-017-0041-5
Zhang, N., Zhong, J., Han, S., Li, Y., Yin, Y., and Li, J. (2016). MicroRNA-378 alleviates cerebral ischemic injury by negatively regulating apoptosis executioner caspase-3. Int. J. Mol. Sci. 17 (9), 1427. doi:10.3390/ijms17091427
Zhang, Q., Chen, C. Y., Yedavalli, V. S., and Jeang, K. T. (2013). NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 4 (1), e00596-12. doi:10.1128/mbio.00596-12
Zhou, L., Pupo, G. M., Gupta, P., Liu, B., Tran, S. L., Rahme, R., et al. (2012). A parallel genome-wide mRNA and microRNA profiling of the frontal cortex of HIV patients with and without HIV-associated dementia shows the role of axon guidance and downstream pathways in HIV-mediated neurodegeneration. BMC Genomics 13 (1), 677. doi:10.1186/1471-2164-13-677
Keywords: antiretroviral therapy, HIV-1, long non-coding RNA, RNA-seq, T-Lymphocytes
Citation: Farsiu N, Ghorbani A, Karbanowicz TP, Mottaghi-Dastjerdi N, Veltri P and Guzzi PH (2024) Discovery of long non-coding RNAs in naïve CD4+ T cells in response to initiating antiretroviral therapy at acute or chronic phase of HIV-1 infection. Acta Virol. 68:11572. doi: 10.3389/av.2024.11572
Received: 12 May 2023; Accepted: 27 March 2024;
Published: 11 April 2024.
Edited by:
Daniel Ruzek, Masaryk University, CzechiaReviewed by:
Ahmad Tahmasebi, Shiraz University, IranDalibor Miklík, Institute of Molecular Genetics (ASCR), Czechia
Copyright © 2024 Farsiu, Ghorbani, Karbanowicz, Mottaghi-Dastjerdi, Veltri and Guzzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abozar Ghorbani, YWJnaG9yYmFueUBhZW9pLm9yZy5pcg==
 Niloofar Farsiu
Niloofar Farsiu Abozar Ghorbani
Abozar Ghorbani Thomas P. Karbanowicz3
Thomas P. Karbanowicz3