Abstract
Coronavirus Disease 2019 (COVID-19) pandemic led to 7 million deaths and more than 770 million confirmed cases worldwide. The Spike glycoprotein (SP) is responsible for recognizing and binding to angiotensin converting enzyme-2 (ACE-2) in the host cell membrane and seems to modulate host cellular signaling pathways. Here, we investigate the effects of SP (stabilized in prefusion conformation) in human umbilical vein endothelial cells (HUVEC-C) lineage on the ACE-2 expression profile and in cell glucose metabolism. Our data indicate that SP binds to ACE-2, is internalized by HUVEC-C cells, and positively modulates ACE-2 expression. In addition, SP alone induces a transient increase in glucose uptake and a decrease in lactate production, characterizing itself as a metabolic regulating protein. The present study is the first to demonstrate that SP induces a slight change in cell metabolism, promotes the overexpression of ACE-2 and its increased availability in the membrane of endothelial cells in a time-dependent fashion.
Introduction
In the 3 years since the Coronavirus disease-19 (COVID-19) pandemic was declared by the World Health Organization (WHO), over 770 million people have been infected worldwide and more than 7 million people died.1 COVID-19 is a respiratory disease characterized by flu-like symptoms that can evolve into severe respiratory distress and death (Lopes-Pacheco et al., 2021). Its causative agent, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has a 26–32 kb long single-stranded positive-sense RNA genome with two open reading frames (ORFs) that generate the polyproteins Pp1a and Pp1ab, containing all the non-structural proteins (Nsp1-16), and multiple subgenomic RNAs that encode the structural proteins (Wang et al., 2020; Lopes-Pacheco et al., 2021). Among them, the Spike glycoprotein (SP) is a key component of the virus particle since it is responsible for recognizing and binding to angiotensin-converting enzyme 2 (ACE-2) in the host cell membrane. It also triggers receptor-mediated endocytosis and promotes viral and endosomal membrane fusion; therefore it is crucial for proper virus entry within the cell (Huang et al., 2020; Walls et al., 2020). In general, besides its role in the cell entry process (Li, 2016), coronaviruses SP modulate host cellular signaling pathways, including i) activation of angiotensin II receptor type 1 (AT1) receptors and the downstream mitogen-activated protein kinase/nuclear factor kappa B (MAPK/NFκB) pathway, ii) enhancement of Janus associated kinase (JAK) and signal transducer and activator transcription factor (STAT) interaction, and iii) induction of mitogen-activated protein kinase kinase (MEK) phosphorylation (Patra et al., 2020; Suzuki et al., 2021; Zhou et al., 2021). Likewise, overexpression of SP is shown to modulate ACE-2 expression and exert a controversial role in SARS-CoVs’ pathophysiology (Kuba et al., 2005; Lei et al., 2020; Patra et al., 2020; Zhou et al., 2021; Singh et al., 2022).
Understanding the effects of SP in different ACE-2-expressing cells is extremely relevant for showing its potential antagonistic role during disease pathogenesis and to anticipate any eventual adverse effect of SP-based vaccines. Endothelial injury and increased vascular permeability are important features of COVID-19 that contribute to vascular damage and organ failure (Lei et al., 2020; Ragab et al., 2020). Recent data indicate that changes in glycolytic metabolism is a key feature for endothelial damage (Icard et al., 2021; Santos et al., 2021). Since endothelial cells express high levels of ACE-2 and are very susceptible to SARS-CoV-2 infection (Ma et al., 2020; Varga et al., 2020), we evaluated the toxicity, ACE-2 expression profile, and metabolic reprogramming of human endothelial cells exposed to recombinant SARS-CoV-2 SP. The present study is the first to demonstrate that SP does not induce toxicity in endothelial cells but promotes ACE-2 overexpression, increases glucose uptake, and reduces intracellular lactate availability. The results presented in the current study contribute to understanding the initial cellular responses triggered by the interaction of SP to its membrane receptor and propose a new role for SP in the pathophysiology of SARS-CoV-2.
Materials and methods
Cell culture and SARS-CoV-2 SP
Human-derived umbilical vein endothelial cells (HUVEC-C, ATCC) were cultivated in F-12 Ham’s medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% PenStrep antibiotics (Gibco). Cells were cultured in a humid atmosphere incubator with 5% CO2 at 37°C. Cultures were propagated every 48 h or until 90% confluence before treatment with recombinant SARS-CoV-2 SP protein following the conditions established for each assay. The trimeric spike protein of SARS-COV-2 stabilized in the prefusion conformation was expressed in stable recombinant HEK293 cells according to Alvim et al. (2022), and was kindly provided by the Cell Culture Engineering Laboratory of COPPE/Universidade Federal do Rio de Janeiro. Detailed information about the SP construct is described by Wrapp and colleagues (Wrapp et al., 2020).
Flow cytometry assays
SARS-CoV-2 spike protein binding and uptake
HUVEC cells were incubated with SP at 50 μg/mL for 30 min at 37°C for the uptake assay and at 4°C for the binding assay. Then, cells were washed twice with PBS 1x (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4 and 2 mM KH2PO4), detached with trypsin for 5 min at 37°C, and fixed with 4% paraformaldehyde. For the uptake assay, after the fixation step, cells were incubated in permeabilization solution (PBS, 0.5% BSA and 0.1% triton x-100). Cells were then incubated with anti-SP monoclonal antibody (GeneTex, GTX632604) for 1 h followed by incubation with Alexa Fluor 488 conjugated-anti-mouse IgG antibody (Invitrogen). Finally, cells were resuspended in PBS 1x prior to analysis using BD FACSCalibur (BD Biosciences).
ACE-2 membrane availability
For measurement of ACE-2 expression, HUVEC cells were treated with SP at either 2.5 or 15 μg/mL for 2, 4, and 16 h before analysis by flow cytometry. Cells were first washed twice with PBS 1x (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4 and 2 mM KH2PO4), detached with trypsin for 5 min at 37°C, fixed with 4% paraformaldehyde, and incubated in blocking solution (PBS, 0.5% BSA). After this step, cells were incubated with anti-ACE-2 monoclonal antibody (Abcam, ab108252) for 1 h followed by incubation with Alexa Fluor 488 conjugated-anti-mouse IgG antibody (Invitrogen). Finally, cells were resuspended in PBS 1x prior to analysis using BD FACSCalibur (BD Biosciences).
HUVEC-C cells were incubated with SP at 50 μg/mL for 30 min at 37°C for the uptake assay and at 4°C for the binding assay. To measure ACE-2 expression, the cells were treated with SP at either 2.5 μg/mL or 15 μg/mL for 2, 4, and 16 h before analysis by flow cytometry. Cells were first washed twice with PBS, detached with trypsin for 5 min at 37°C, fixed with 4% paraformaldehyde, and incubated in permeabilization solution (PBS, 0.5% BSA, and 0.1% triton x-100). After this step, cells were incubated with either anti-ACE-2 monoclonal antibody (Abcam, ab108252) or anti-S monoclonal antibody (GeneTex, GTX632604) for 1 h followed by incubation with Alexa Fluor 488 conjugated-anti-mouse IgG antibody (Invitrogen). Finally, cells were resuspended in PBS 1x (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4) before analysis using BD FACSCalibur (BD Biosciences).
Cell death
Cell viability was determined by labeling the cells with Annexin-V and Propidium iodide (PI) as recommended by the manufacturer. After incubation, cells were washed twice with PBS and detached with trypsin-EDTA solution for 5 min at 37°C. Then DMEM medium supplemented with 10% fetal bovine serum was added, and the cells were centrifuged. About 106 cells were resuspended in buffer solution (BD PharMingen), and Annexin-V and PI were added and incubated in the dark for 15 min. Cells were acquired in a FACSCalibur (BD Biosciences) flow cytometer. Data from both experiments were analyzed using the FlowJO software.
MTT assays
HUVEC-C cells were treated with SP (2.5, 7.5, and 15 μg/mL) for 16 h or 15 μg/mL for 2, 4, and 16 h at 37°C with 5% CO2 before assessment of cell viability by MTT colorimetric assay. After treatments, culture media were discarded, and fresh culture medium containing 5 mM MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide—Invitrogen] was added into each well. The cells were incubated at 37°C with 5% CO2 for 4 h in a humid atmosphere. DMSO was used to dissolve MTT crystals, and the absorbance was measured at 540 nm. Each condition was analyzed in triplicate, and the final data resulted from three independent experiments.
Glucose uptake and lactate measures
Glucose uptake and lactate were measured with Promega Corporation luminescent kits (Glucose Uptake-Glo™ Assay and Lactate-Glo™ Assay) following the manufacturer’s instructions. HUVEC-C cells grown in 96-well plates were treated with 15 μg/mL of the recombinant SARS-CoV-2 spike protein for 1–4 h, and the luminescence readings were performed in a SpectraMax M5 (Molecular Devices) microplate reader.
RNA isolation and quantitative real-time PCR
Total RNA extraction from HUVEC-C cells treated with 15 μg/mL SP for 2 and 4 h was performed using PureLink™ RNA Mini Kit (Invitrogen, ThermoFisher Scientific). Total RNA was conversed into cDNA by using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). The reaction was carried out in a QuantStudio™ 3 Real-Time PCR System (ThermoFisher Scientific) using the Applied Biosystems Power SYBR® Green RT-PCR Reagents kit (ThermoFisher Scientific). All reactions were made in triplicate. Specific primers for the ACE-2 gene are:foward: 5′- GGGATCAGAGATCGGAAGAAGAAA—3′ and reverse: 5′- AGGAGGTCTGAACATCATCAGTG—3′. Human β-actin gene (ACTB) was used for normalization.
Statistical analysis
The data satisfied parametric assumptions and were expressed as the mean and SD. Repeat-measure analysis of variance (RM-ANOVA) followed by Dunnet’s Multiple Comparison Test was then conducted for statistical analysis, and statistical significance was established at p < 0.05. All tests were carried out in GraphPad Prism version 9.1.0 (GraphPad Software, San Diego, CA). FlowJo software v10.9 was used for flow cytometry analysis. At least three independent experiments were performed for each assay.
Results
SARS-CoV-2 SP binds to and is internalized by HUVEC-C cells but does not induce significant cell toxicity
We first evaluated whether exposure to exogenous recombinant SP alone would result in membrane binding and uptake by HUVEC-C cells. As shown in Figure 1A, exogenous SP not only bound to membrane receptors, but it was also internalized by these cells. Previous literature shows that SP reduces around 70% Caco-2 cell viability after 24 h incubation (Corpetti et al., 2021), therefore, we assessed whether this toxicity would also occur in HUVEC-C cells. Incubation with 15 μg/mL of SP for up to 16 h reduced HUVEC-C cell viability in around 12% of cells, resulting in no statistical difference compared to non-treated cells (Figure 1B). This indicates that endothelial cells are more resistant to SP-induced toxicity. As some studies have demonstrated that SP overexpression induces cell death (Barhoumi et al., 2021; Li et al., 2021), this process was also investigated in HUVEC-C cells exposed to SP. Interestingly, significant induction of either early/late apoptosis or necrosis compared to non-treated cells after 2 or 4 h exposure to SP (Figures 1C, D) was not observed, suggesting that binding and internalization of SP is not enough to induce cell death in a short period of time.
FIGURE 1
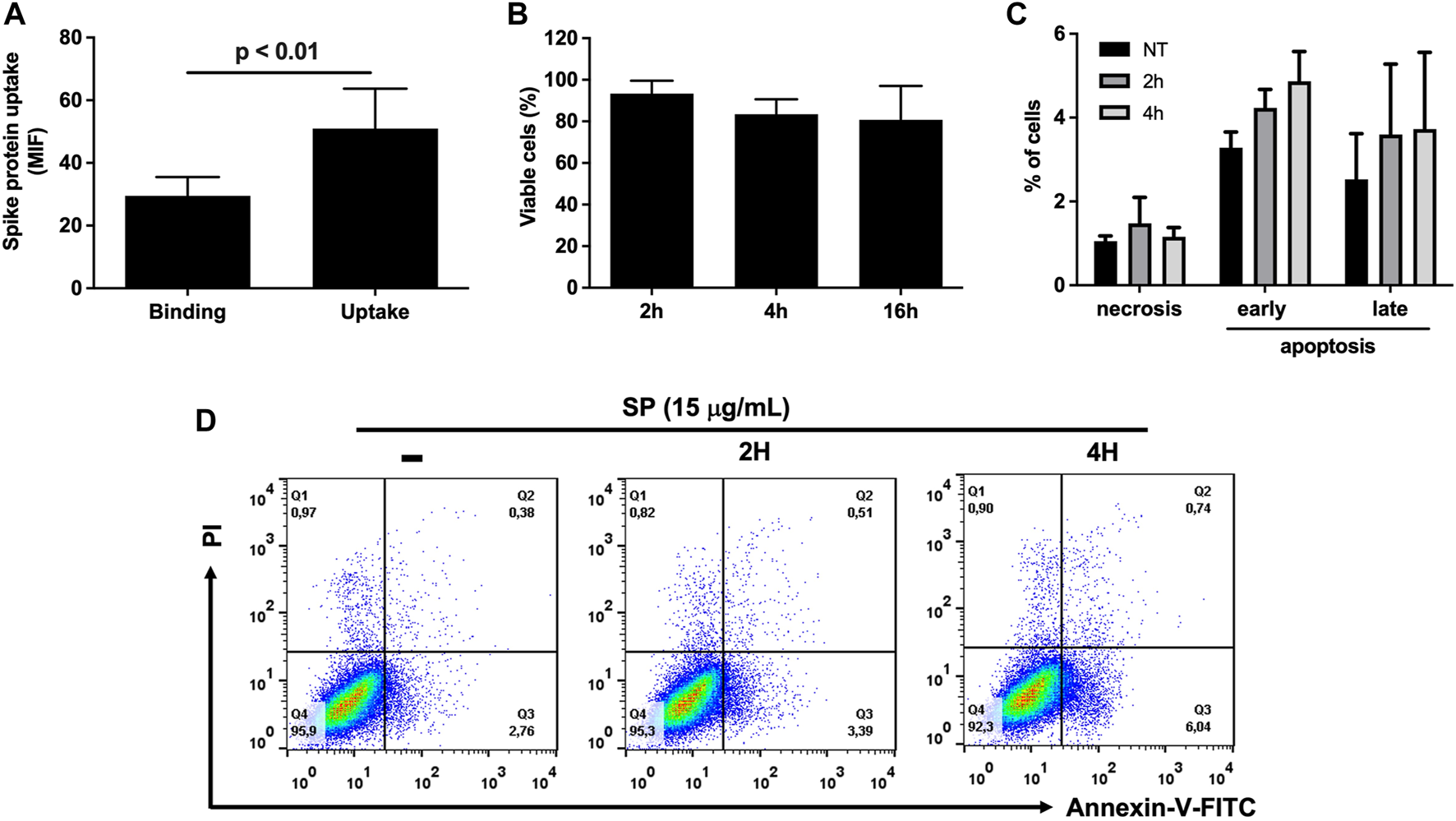
Effects of SARS-CoV-2 SP in HUVEC-C cells. (A) Binding and uptake of SP by HUVEC-C cells, at a concentration of 50 μg/mL, were assessed by flow cytometry. MIF: Mean Fluorescence Intensity. (B) Percentage of cell viability after treatment with 15 μg/mL SP for 2, 4, and 16 h, assessed by MTT colorimetric assay. (C) Percentage of cell death (necrosis, early/late apoptosis) after HUVEC-C cells were exposed to SP for 2 h and 4 h, assessed by Annexin-PI staining in flow cytometry. (D) Flow cytometry dot blot of HUVEC-C cells exposed to 15 μg/mL SP. NT: non-treated cells.
SARS-CoV-2 SP increases ACE-2 expression and alters glucose metabolism
As most of exposed SP was internalized by HUVEC-C cells, and considering that ACE-2 is its major receptor, we investigated whether membrane ACE-2 expression levels were modulated by SP. ACE-2 expression in membranes of HUVEC-C cells after 2 h of SP exposure was significantly increased compared to non-treated cells (Figure 2A, p < 0.05). This phenomenon was not observed after 4 h of exposure. To understand whether this increment in ACE-2 levels was due to its increased expression or its re-localization to cell membranes, we performed a qPCR of HUVEC-C cells exposed to 15 μg/mL of SP for 2 and 4 h. As shown in Figure 2B, there was a significant increase in ACE-2 gene expression after 2 h of SP exposure compared to non-treated cells (p < 0.05), but it was not observed after 4 h, corroborating the previous result and suggesting that SP triggers an internal signal that culminates in the time-dependent ACE-2 overexpression.
FIGURE 2
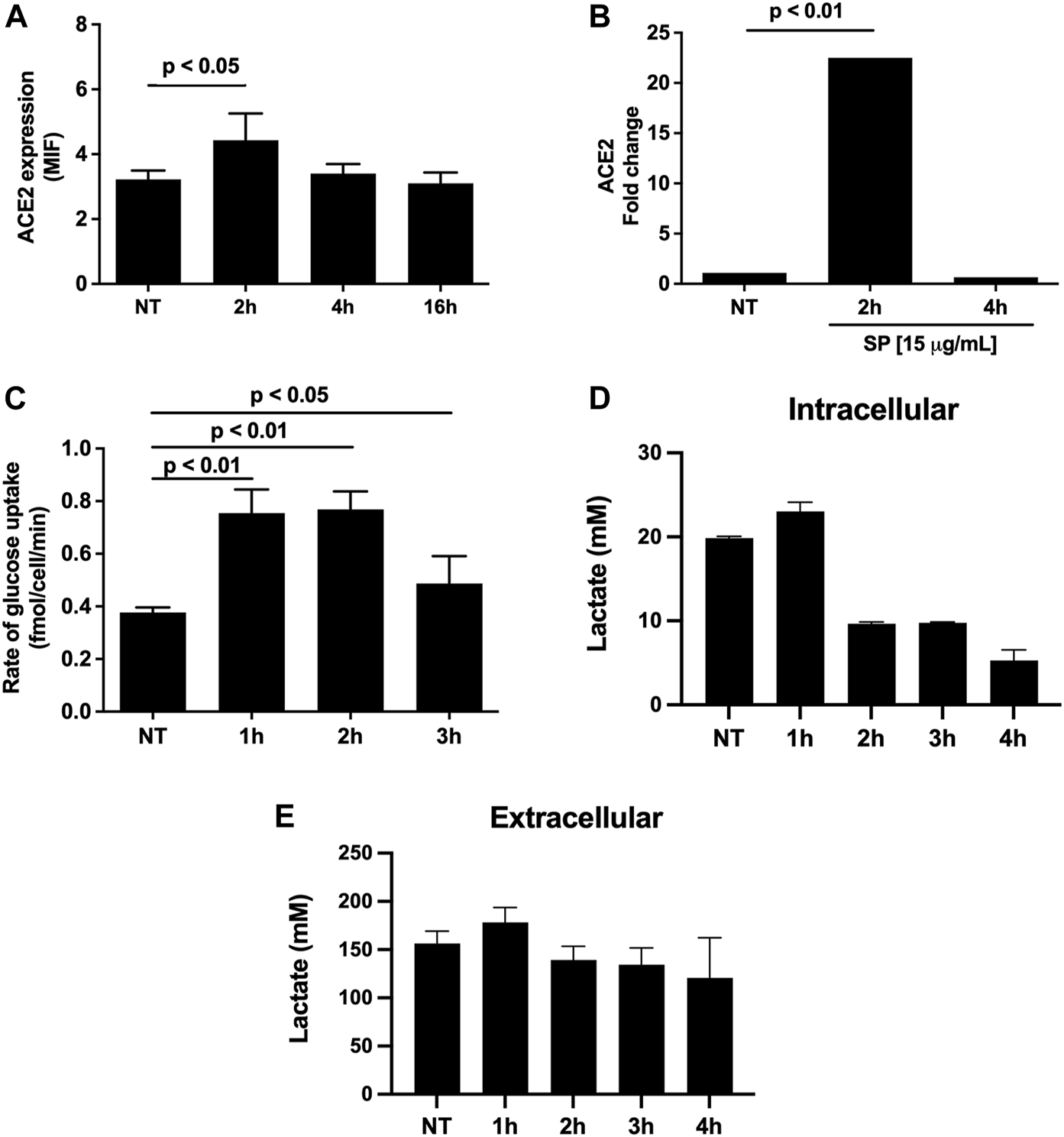
Metabolic regulation mediated by SP in HUVEC-C cells. ACE2 availability (A) and expression (B) after 2, 4, and 16 h of exposure to 15 μg/mL SARS-CoV-2 SP. (C) Rate of glucose uptake in HUVEC-C cells after 1, 2, and 3 h of exposure to 15 μg/mL of SARS-CoV-2 SP. (D) Intracellular and (E) extracellular lactate measures in HUVEC-C cells after 1, 2, 3, and 4 h of exposure to 15 μg/mL of SARS-CoV-2 recombinant Spike protein. Statistical analyses were performed by using RM-Anova. These are the means of three independent experiments. NT = non-treated cells. MIF: Mean Fluorescence Intensity.
SARS-CoV-2 infection induces important changes in the glycolytic pathway, including the promotion of the Warburg effect in endothelial cells, which is associated with endothelial cell dysfunction (Icard et al., 2021; Santos et al., 2021). Therefore, we assessed whether SP could promote any change in glucose uptake as indicative of its role in aerobic glycolysis promoted by SARS-CoV-2. Interestingly, there was a significant increase in glucose uptake after 1 and 2 h (p < 0.05) of SP exposure compared to non-treated cells, but this effect was not observed after 3 h of SP incubation (Figure 2C). However, the glucose uptake increment did not reflect an increase in lactate production, since SP reduces intracellular lactate production after 2 h of exposure (Figure 2D) without increasing lactate release (Figure 2E). This indicates that internalized glucose should be moving toward a tricarboxylic acid (TCA) cycle instead of aerobic glycolysis. These data suggest that SP can modulate cell glycolytic metabolism by increasing glucose uptake and probably by driving it to the oxidative phosphorylation pathway in the first moments after binding to ACE-2/internalization by HUVEC-C cells.
Discussion
Viruses are known to modulate several cellular processes, including protein expression and subcellular localization of key molecules, to allow a successful infection. Mapping which viral protein contributes to this process is of utmost importance to understanding the disease pathophysiology and providing new pharmacological targets for future antiviral development. In the case of SARS-CoV-2, literature has focused on depicting the role of nonstructural proteins during the metabolic changes induced by virus infection. In this study, we show that SARS-CoV-2 SP, a structural protein, can modulate ACE-2 availability in the human endothelial cell membrane and induce an increase in glucose uptake and a reduction of intracellular lactate production.
Several COVID-19 patients exhibited endothelitis (Varga et al., 2020) and the deregulation of vascular homeostasis is an important feature of COVID-19 severity, making these cells an important model to investigate the disease pathophysiology (Ackermann et al., 2020; Lopes-Pacheco et al., 2021). It was recently demonstrated that SARS-CoV-2 has a tropism for endothelial cells since they express not only ACE-2 but also TMPRSS2, which are essential for the virus entry [9,15]. Although these two proteins are present in HUVEC-C cells (data not shown), the SP used here consists of a full-length ectodomain stabilized in the prefusion trimeric conformation (Alvim et al., 2022) in which the furin cleavage site was removed. Thus, the effects observed here are results from internalized SP by endocytic process triggered by SP:ACE-2 interaction.
When the viral infection is ongoing, each stage of the viral life cycle can generate different cellular responses, including changes in cell metabolism to favor the replication (Goodwin et al., 2015). Interaction between cell receptors and virus ligands alone can be responsible for initiating, mediating and controling these responses (Moolamalla et al., 2021; Umar et al., 2022). SARS-CoV-2 can interfere with several intracellular pathways in host cells, including glucose metabolism (Shen and Wang, 2021; Martínez-Gómez et al., 2022). It was already described that SARS-CoV-2 induces the production and secretion of lactate, pyruvate, pyruvate kinase, and lactate dehydrogenase (LDH), which are all related to glucose metabolism, turning infected cells highly glycolytic, which is a common feature in COVID-19 patients (Codo et al., 2020; Shen and Wang, 2021). Our data show that SP can modulate cell metabolism by promoting higher glucose uptake and probably driving energy metabolism to oxidative phosphorylation instead of aerobic glycolysis, confirming its metabolic regulatory role. Although our results indicate, at first glance, an opposite direction from the literature, it is worth noting that we are studying a single viral protein in an in vitro model that simulates a very early event of the infection process. During virus infection, several structural and nonstructural proteins are being expressed and the outcome is a balance of all their activities. Notwithstanding, depicting alternative functions of virus proteins is very important in the context of virus biology.
ACE-2 levels in cell membrane seem to be crucial for proper SARS-CoV-2 infection since its availability in cell membranes directly impacts the success of virus infection (Kuba et al., 2005). However, SP has already been linked to ACE-2 downregulation in lung and endothelial cells, which was correlated to the development of lung injury (Kuba et al., 2005; Glowacka et al., 2010) and the worst prognosis of COVID-19 (Lei et al., 2020), respectively. In all situations, the ACE-2 downregulation occurred after longer periods of SP intracellular overexpression, which reflects a later stage of the virus life cycle. Our data demonstrate that SP binding to ACE-2 and its internalization induces the upregulation of ACE-2 gene expression and its increased availability in cell membranes in a relatively short period of time, which may be an important feature to guarantee proper cell infection and minimize cellular antiviral responses during initial moments of SARS-CoV-2 infection.
Overall, our findings provide the first pieces of evidence that the SARS-CoV-2 SP promotes the modulation of the endothelial cell metabolism and increases ACE-2 availability on the host cell membrane, in a time-dependent fashion, without inducing cell toxicity. Further studies are necessary to detail the mechanisms displayed by SP involved in metabolism modulation.
Statements
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors upon request, without undue reservation.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
MC: conceptualization, methodology, resources, investigation, formal analysis, writing–original draft; LC: methodology, resources, investigation; DT: methodology, investigation, formal analysis; RS-A.: methodology, investigation; PR: conceptualization, writing–reviewing and editing; RM-B: conceptualization, funding acquisition, writing–reviewing and editing; GL: conceptualization, funding acquisition, writing–reviewing and editing; CC-N: conceptualization, funding acquisition, writing–reviewing and editing; SL: conceptualization, funding acquisition, supervision; writing–reviewing and editing; DA: conceptualization, methodology, formal analysis, supervision, project administration, funding acquisition, writing–reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
We would like to thank the financial support of Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES (code 001) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro—FAPERJ (E-26/210.403/2022, E-26/201.303/2022, and E-26/211.285/2021).
Acknowledgments
We thank the Cell Culture Engineering Laboratory of COPPE/UFRJ—Federal University of Rio de Janeiro (Brazil) for providing the trimeric SP of SARS-COV-2.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
1
Ackermann M. Verleden S. E. Kuehnel M. Haverich A. Welte T. Laenger F. et al (2020). Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med.383, 120–128. 10.1056/NEJMoa2015432
2
Alvim R. G. F. Lima T. M. Rodrigues D. A. S. Marsili F. F. Bozza V. B. T. Higa L. M. et al (2022). From a recombinant key antigen to an accurate, affordable serological test: lessons learnt from COVID-19 for future pandemics. Biochem. Eng. J.186, 108537. 10.1016/j.bej.2022.108537
3
Barhoumi T. Alghanem B. Shaibah H. Mansour F. A. Alamri H. S. Akiel M. A. et al (2021). SARS-CoV-2 coronavirus spike protein-induced apoptosis, inflammatory, and oxidative stress responses in THP-1-like-macrophages: potential role of angiotensin-converting enzyme inhibitor (perindopril). Front. Immunol.12, 728896. 10.3389/fimmu.2021.728896
4
Codo A. C. Davanzo G. G. Monteiro L. de B. de Souza G. F. Muraro S. P. Virgilio-da-Silva J. V. et al (2020). Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab.32, 437–446.e5. 10.1016/j.cmet.2020.07.007
5
Corpetti C. Del Re A. Seguella L. Palenca I. Rurgo S. De Conno B. et al (2021). Cannabidiol inhibits SARS-Cov-2 spike (S) protein-induced cytotoxicity and inflammation through a PPARγ-dependent TLR4/NLRP3/Caspase-1 signaling suppression in Caco-2 cell line. Phytother. Res.35, 6893–6903. 10.1002/ptr.7302
6
Glowacka I. Bertram S. Herzog P. Pfefferle S. Steffen I. Muench M. O. et al (2010). Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol.84, 1198–1205. 10.1128/JVI.01248-09
7
Goodwin C. M. Xu S. Munger J. (2015). Stealing the keys to the kitchen: viral manipulation of the host cell metabolic network. Trends Microbiol.23, 789–798. 10.1016/j.tim.2015.08.007
8
Huang Y. Yang C. Xu X.-F. Xu W. Liu S.-W. (2020). Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin.41, 1141–1149. 10.1038/s41401-020-0485-4
9
Icard P. Lincet H. Wu Z. Coquerel A. Forgez P. Alifano M. et al (2021). The key role of Warburg effect in SARS-CoV-2 replication and associated inflammatory response. Biochimie180, 169–177. 10.1016/j.biochi.2020.11.010
10
Kuba K. Imai Y. Rao S. Gao H. Guo F. Guan B. et al (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med.11, 875–879. 10.1038/nm1267
11
Lei Y. Zhang J. Schiavon C. R. He M. Chen L. Shen H. et al (2020). SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE2. bioRxiv, 2020.12.04.409144. 10.1101/2020.12.04.409144
12
Li F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol.3, 237–261. 10.1146/annurev-virology-110615-042301
13
Li F. Li J. Wang P.-H. Yang N. Huang J. Ou J. et al (2021). SARS-CoV-2 spike promotes inflammation and apoptosis through autophagy by ROS-suppressed PI3K/AKT/mTOR signaling. Biochim. Biophys. Acta Mol. Basis Dis.1867, 166260. 10.1016/j.bbadis.2021.166260
14
Lopes-Pacheco M. Silva P. L. Cruz F. F. Battaglini D. Robba C. Pelosi P. et al (2021). Pathogenesis of multiple organ injury in COVID-19 and potential therapeutic strategies. Front. Physiol.12, 593223–23. 10.3389/fphys.2021.593223
15
Ma D. Chen C.-B. Jhanji V. Xu C. Yuan X.-L. Liang J.-J. et al (2020). Expression of SARS-CoV-2 receptor ACE2 and TMPRSS2 in human primary conjunctival and pterygium cell lines and in mouse cornea. Eye (Lond)34, 1212–1219. 10.1038/s41433-020-0939-4
16
Martínez-Gómez L. E. Ibarra-González I. Fernández-Lainez C. Tusie T. Moreno-Macías H. Martinez-Armenta C. et al (2022). Metabolic reprogramming in SARS-CoV-2 infection impacts the outcome of COVID-19 patients. Front. Immunol.13, 936106. 10.3389/fimmu.2022.936106
17
Moolamalla S. T. R. Balasubramanian R. Chauhan R. Priyakumar U. D. Vinod P. K. (2021). Host metabolic reprogramming in response to SARS-CoV-2 infection: a systems biology approach. Microb. Pathog.158, 105114. 10.1016/j.micpath.2021.105114
18
Patra T. Meyer K. Geerling L. Isbell T. S. Hoft D. F. Brien J. et al (2020). SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PloS Pathog.16, e1009128. 10.1371/journal.ppat.1009128
19
Ragab D. Salah Eldin H. Taeimah M. Khattab R. Salem R. (2020). The COVID-19 cytokine storm; what we know so far. Front. Immunol.11, 1446. 10.3389/fimmu.2020.01446
20
Santos A. F. Póvoa P. Paixão P. Mendonça A. Taborda-Barata L. (2021). Changes in glycolytic pathway in SARS-COV 2 infection and their importance in understanding the severity of COVID-19. Front. Chem.9, 685196. 10.3389/fchem.2021.685196
21
Shen T. Wang T. (2021). Metabolic reprogramming in COVID-19. Int. J. Mol. Sci.22, 11475. 10.3390/ijms222111475
22
Singh R. D. Barry M. A. Croatt A. J. Ackerman A. W. Grande J. P. Diaz R. M. et al (2022). The spike protein of SARS-CoV-2 induces heme oxygenase-1: pathophysiologic implications. Biochim. Biophys. Acta Mol. Basis Dis.1868, 166322. 10.1016/j.bbadis.2021.166322
23
Suzuki Y. J. Nikolaienko S. I. Dibrova V. A. Dibrova Y. V. Vasylyk V. M. Novikov M. Y. et al (2021). SARS-CoV-2 spike protein-mediated cell signaling in lung vascular cells. Vasc. Pharmacol.137, 106823. 10.1016/j.vph.2020.106823
24
Umar S. Palasiewicz K. Meyer A. Kumar P. Prabhakar B. S. Volin M. V. et al (2022). Inhibition of IRAK4 dysregulates SARS-CoV-2 spike protein-induced macrophage inflammatory and glycolytic reprogramming. Cell Mol. Life Sci.79, 301. 10.1007/s00018-022-04329-8
25
Varga Z. Flammer A. J. Steiger P. Haberecker M. Andermatt R. Zinkernagel A. S. et al (2020). Endothelial cell infection and endotheliitis in COVID-19. Lancet395, 1417–1418. 10.1016/S0140-6736(20)30937-5
26
Walls A. C. Park Y.-J. Tortorici M. A. Wall A. McGuire A. T. Veesler D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell181, 281–292.e6. 10.1016/j.cell.2020.02.058
27
Wang M. Y. Zhao R. Gao L. J. Gao X. F. Wang D. P. Cao J. M. (2020). SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell Infect. Microbiol.10, 587269. 10.3389/fcimb.2020.587269
28
Wrapp D. Wang N. Corbett K. S. Goldsmith J. A. Hsieh C. L. Abiona O. et al (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science367 (6483), 1260–1263. 10.1126/science.abb2507
29
Zhou Y. Wang M. Li Y. Wang P. Zhao P. Yang Z. et al (2021). SARS-CoV-2 Spike protein enhances ACE2 expression via facilitating Interferon effects in bronchial epithelium. Immunol. Lett.237, 33–41. 10.1016/j.imlet.2021.06.008
Summary
Keywords
COVID-19, SARS-CoV-2, spike protein, endothelial cells, HUVEC-C cells, metabolic reprogramming
Citation
Campos MF, Constant LEC, Teixeira DE, Silva-Aguiar RP, Rocco PRM, Mohana-Borges R, Leitão GG, Caruso-Neves C, Leitão SG and Allonso D (2024) SARS-CoV-2 spike protein increases angiotensin converting enzyme-2 expression and promotes an increase in glucose uptake in endothelial cells. Acta Virol. 68:12136. doi: 10.3389/av.2024.12136
Received
27 September 2023
Accepted
20 February 2024
Published
04 March 2024
Volume
68 - 2024
Edited by
Amir Ghorbani, National Institutes of Health (NIH), United States
Reviewed by
Brandt D. Pence, University of Memphis, United States
Wen Meng, University of Pittsburgh, United States
Updates
Copyright
© 2024 Campos, Constant, Teixeira, Silva-Aguiar, Rocco, Mohana-Borges, Leitão, Caruso-Neves, Leitão and Allonso.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diego Allonso, diegoallonso@pharma.ufrj.br; Suzana G. Leitão, sgleitao@pharma.ufrj.br
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.