Abstract
Background:
Currently, there are approximately 240 million individuals worldwide suffering from chronic hepatitis B infection, yet a definitive cure remains elusive. In the context of pharmaceutical interventions for this patient population, the potential issue of Hepatitis B virus (HBV) reactivation must be taken into consideration. Nutlin-3 is a small molecule of imidazoline analog, which inhibits tumor growth by suppressing the interaction between human double minute 2 (HDM2) and p53. However, whether it affects HBV replication is not clear.
Methods:
HepG2.2.15 cells were used to assess the impact of Nutlin-3 on HBV replication. The expression of Hepatitis B surface antigen (HBsAg) and Hepatitis B e antigen (HBeAg) was detected by ELISA. The expression of pregenomic RNA (pgRNA) and core-DNA was detected by RT-qPCR. The activation of relevant signaling pathways was assessed through Western blotting and confocal microscopy detection of the expression of associated proteins.
Results:
We found that Nutlin-3 significantly enhances the expression of HBsAg and HBeAg, and it also promotes the expression of pgRNA and Core-DNA. Nutlin-3 promotes HBV replication independent of the HDM2. Instead, it activates the autophagy pathway through p53-mediated mTOR inhibition to promote HBV replication.
Conclusion:
When using Nutlin-3 for cancer therapy, it’s important to be aware of the potential side effect of promoting HBV replication in individuals with chronic hepatitis B infection.
Introduction
Hepatitis B virus (HBV) is a DNA virus which may cause acute or chronic hepatitis. Globally, it is estimated that over 2 billion people are infected with the HBV, with around 240 million individuals suffering from long-term consequences of chronic HBV infection (Yardeni et al., 2023).
Current therapeutic approaches for chronic hepatitis B aim to achieve sustained viral suppression, alleviate liver inflammation, and prevent disease progression. The mainstay of treatment involves antiviral medications, such as nucleos(t)ide analogs (NAs) or pegylated interferon-alpha (Fung et al., 2022). NAs, including drugs like entecavir and tenofovir, work by inhibiting viral DNA replication, thereby suppressing viral load. While these medications effectively control viral replication, they often necessitate long-term use, and discontinuation may lead to viral rebound. Pegylated interferon-alpha, on the other hand, modulates the immune response to the virus. It has a finite treatment duration but is associated with side effects that may limit its tolerability for some patients (Block et al., 2016).
Nutlin-3 is a small molecule of imidazoline analog, which inhibits the HDM2-p53 interaction (Dey et al., 2007). p53 serves as a vital tumor suppressor, playing a crucial role in maintaining cellular genetic stability and preventing the development of cancer (Levine, 1997). p53 mutations or inactivation usually occur in around 50% of human cancers (Vogelstein et al., 2000). HDM2, an E3 ubiquitin-protein ligase, targets p53 for its proteasome-mediated degradation (Haupt et al., 1997). In turn, p53 also promotes HDM2 gene expression through binding to its promoter (Psatha et al., 2023). Therefore, Nutlin-3 concurrently upregulates the expression of both HDM2 and p53 through disrupting the interaction between HDM2 and p53. Nutlin-3 has been extensively researched and recognized as a promising anticancer drug candidate (Supiot et al., 2008; Tabe et al., 2009; Vassilev et al., 2004; Voon et al., 2015; Yee-Lin et al., 2018; Zheng et al., 2014). HDM2 has been reported to enhance the NEDDylation of Hepatitis X protein (HBx) to increase its stability, promoting HBV replication (Liu et al., 2017). p53 has been found to possess the capability to inhibit HBV transcription (Ori et al., 1998). Therefore, Nutlin-3 may affect HBV replication, but currently, there are no reported findings.
In this study, we found that Nutlin-3 significantly promote HBV replication in HepG2.2.15 cell. Nutlin-3 promotes HBV replication independent of HDM2. Instead, it activates the autophagy pathway through p53-mediated mTOR inhibition to promote HBV replication. This study indicates that when using Nutlin-3 for cancer therapy, the potential side effect of promoting HBV replication in individuals with chronic hepatitis B infection may be considered.
Materials and methods
Reagents
Nutlin-3 was purchased from Beyotime Biotechnology (Shanghai, China). Dulbecco’s Modified Eagle Medium (DMEM medium) was purchased from Invitrogen (CA, United States). G418 was purchased from Shanghai Sangon Bioengineering Co., Ltd. Penicillin and streptomycin were purchased from Solaribio (Beijing, China). Fetal bovine serum was purchased from Sigma-Aldrich (St. Louis, MO, United States). CCK8 detection kit was purchased from EnoGene (Nanjing, China). ExFect Transfection Reagent was purchased from Vazyme (Nanjing, China).
HBsAg and HBeAg Elisa kit was purchased from Beijing Wantai Biological Pharmaceutical Co., Ltd. HDM2 (ab259265) and p53 primary antibody (ab26) were purchased from Abcam (Cambridge, MA, United States), β-actin antibody (D191048-0100) was purchased from Shanghai Sangon Bioengineering Co., Ltd. Antibodies of LC3B (A19665), p-4E-BP1 (AP0030) were purchased from ABclonal (Wuhan, China). Antibodies of p62 (18420 -1-AP), p-S6K1 (9234T), S6K1 (9202S) were obtained from proteintech (Wuhan, China). ECL (enhanced chemiluminescence) detection kit was purchased from Affinity Bios ciences (OH, United States). All other chemicals used are of analytical grade unless otherwise noted.
Cell culture
HepG2.2.15 cells are a subline of the HepG2 liver carcinoma cell line, stably transfected with the HBV genome, used extensively to study HBV replication and drug effects (Sells et al., 1987). The cells were cultured at 37°C in a humidified atmosphere containing 95% air and 5% CO2. HepG2.2.15 cells were routinely cultured in DMEM medium, supplemented with 10% FBS, 100 U/mL penicillin, 100 U/mL streptomycin, and 400 μg/mL G418. G418 were withdrawn before the cells were plated.
Lentivirus production and infection
The lentivirus expressing the mCherry-EGFP-LC3B fusion protein (Lenti-mCherry-EGFP-LC3B) was obtained from Beyotime (Shanghai, China). This lentivirus was used to infect HepG2.2.15 cells, which were treated with 8 μg/mL of polybrene to facilitate viral entry. Two days post-infection, the cells were selected with 8 μg/mL puromycin for 7 days to establish a stable cell line overexpressing mCherry-EGFP-LC3B. To construct the shRNA lentiviral plasmids, shRNA primer pairs were ligated into a lentiviral vector. Once sequence accuracy was confirmed, the plasmids were co-transfected with lentiviral packaging plasmids into HEK293T cells using the ExFect Transfection Reagent according to the manufacturer’s instructions. Forty-eight hours post-transfection, the medium containing the virus was collected, centrifuged, and filtered to remove cellular debris. The purified viral medium was then used to infect HepG2 cells. The subsequent process for establishing a stable cell line was similar to that used for the mCherry-EGFP-LC3B cell line. The primer sequences for shHDM2 are as follows: Forward: 5ʹ-AATTGATTCCAGAGAGTCATGTGTTCTCGAGAACACATGACTCTCTGGAATCtttttt-3ʹ and Reverse: 5′-GATTCCAGAGAGTC-ATGTGTTCTCGAGAACACATGACTCTCTGGAATC-3′. The primer sequences for shTP53 are: Forward: 5ʹ-AATTGTCCAGA-TGAAGCTCCCAGAACTCGAGTTCTGGGAGCTTCATCTGGACtttttt-3ʹ and Reverse: 5′-GTCCAGATGAAGCTCCCAGAACTC-GAGTTCTGGGAGCTTCATCTGGAC-3′.
Cell viability
HepG2.2.15 cells were seeded in 96-well plates to a cell density of 70%. Then, the medium was removed and replaced with fresh medium containing different concentrations of Nutlin-3 (0.1, 1, 10 μM) for 48 h. Next, the supernatant was collected and 100 μL of CCK8 working solution was added to each well. After incubation for 40 min, cellular activity was determined by analysis of absorbance at 450 nm.
Determination of hepatitis B virus antigen
HBsAg and HBeAg kits (Beijing Wantai Biopharmaceutical Co., Ltd.) were used to detect the levels of HBsAg and HBeAg. Briefly, HepG2.2.15 cells were seeded in 96-well plates and incubated overnight. Then, the medium was replaced with different concentrations of Nutlin-3 (0.1, 1, and 10 μM). The medium was harvested at 48 h. After centrifugation to remove cell debris, the supernatant was collected for detection of HBsAg and HBeAg. Experiments were performed using an enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions.
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were extracted from HepG 2.2.15 cells using EZ-Press RNA Purification Kit. 1 μg of RNA was reverse transcribed to cDNA according to the manufacturer’s protocol. The mRNA levels of target genes were measured by RT-qPCR using SYBR Q-PCR mix (Wuhan, China). The thermal cycling conditions were as follows: pre-denaturation at 95°C for 10 min, denaturation at 95°C for 10 s, followed by 40 cycles of annealing/extension at 60°C for 30 s. After that, we normalized the expression level of the target gene to that of β-actin. The fold change is the 2−ΔΔCT method.
Primers for RT-qPCR,
HDM2-F: GAATCATCGGACTCAGGTACATC
HDM2-R: TCTGTCTCACTAATTGCTCTCCT
TP53-F: CAGCACATGACGGAGGTTGT
TP53-R: TCATCCAAATACTCCACACGC
Total RNA-F: ACGTCCTTTGTTTACGTCCCGT
Total RNA-R: CCCAGCTCCTCCCAGTCCTTAA
pgRNA-F: TCATCCAAATACTCCACACGC
pgRNA-R: GAGGGAGTTCTTCTTCTAGG
β-actin-F: CATGTACGTTGCTATCCAGGC
β-actin-R: CTCCTTAATGTCACGCACGAT
Extraction and detection of HBV Core-DNA
HBV core-associated DNA was extracted utilizing a previously established methodology (Ma et al., 2020). In brief, cells were lysed in NP-40 lysis buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 50 mM NaCl, and 0.5% NP-40) for 10 min at 37°C. Following centrifugation at 4,000×g for 5 min, the lysates were carefully decanted, and the resulting supernatants were transferred to new centrifuge tubes. Micrococcal nuclease was introduced, and the tubes were incubated at 37°C for 2 h to facilitate the digestion of plasmid DNAs. Subsequently, the enzymes were deactivated by adding EDTA (40 mM). The isolation of core-associated DNA was accomplished through a series of steps, including pronase digestion, phenol-chloroform extraction, and ethanol precipitation. Detection of HBV core-associated DNA was performed using a Hepatitis B virus nucleic acid quantitative detection kit following the manufacturer’s instructions.
Western Blot analysis
Total proteins were extracted from HepG2.2.15 cells using RIPA buffer supplemented with a protease inhibitor. Subsequently, the proteins underwent separation on 10 or 12% SDS-PAGE gels and were electrotransferred to polyvinylidene fluoride (PVDF) membranes. These membranes were then blocked with 5% non-fat milk for 1 h at room temperature, followed by overnight incubation at 4°C with primary antibodies targeting HDM2 (1:1,000), p53 (1:1,000), p-4E-BP1 (1:1,000), S6K1 (1:1,000) and p-S6K1 (1:1,000), with β-actin (1:2,000) used as a control. The following day, the membranes were incubated with a secondary goat anti-rabbit IgG (H+L) or anti-mouse IgG (H+L) antibodies for an hour at room temperature. Detection of the protein bands was achieved using ECL detection kit. Band intensities were analyzed with Image J software.
Confocal microscopy
HepG2 cells stably expressing mCherry-GFP-LC3B were seeded onto laser confocal dishes and cultured overnight. Subsequently, Nutlin-3 was added, with DMSO serving as the control. After 48 h, images were captured using a laser confocal microscope.
Statistical analysis
All quantitative data are presented as mean ± standard deviation (SD) and were compared using Student’s t-test or one-way analysis of variance (ANOVA). p < 0.05 was considered statistically significant.
Results
Nutlin-3 promotes HBV replication
To determine whether Nutlin-3 affect HBV replication, Hep2.2.15 cells were treated with different concentrations of Nutlin-3. Given that Nutlin-3 is dissolved in DMSO, an equal volume of DMSO (0.1%) served as the control in these experiments. The level of HBsAg and HBeAg in cell supernatant was measured by ELISA. As Figures 1A, B shown, Nutlin-3 promoted HBsAg and HBeAg secretion in a dose-dependent manner. Even at a concentration of 10 μM, Nutlin-3 did not affect the proliferation of HepG2.2.15 cells (Figure 1C). HBV produces five unique RNA transcripts: 3.5 kb of both pgRNA and precore RNA (pcRNA), 2.4 kb preS1 mRNA, 2.1 kb preS2/S mRNA, and 0.7 kb X mRNA, essential for its replication and protein production processes. The expression of pgRNA and HBV total RNA was detected by RT-qPCR. The results indicate that Nutlin-3 significantly promotes the expression of pgRNA, but does not have a significant impact on total RNA expression (Figures 1D, E). pgRNA plays a vital role in the HBV replication cycle, serving as both the template for reverse transcription into viral DNA and as mRNA for viral protein synthesis. Nutlin-3 significantly enhances pgRNA expression, suggesting it may also markedly promote the synthesis of HBV DNA. Consequently, we investigated the expression of HBV Core-DNA and discovered that Nutlin-3 substantially increases its expression at a concentration of 10 μM (Figure 1F). These results suggest that Nutlin-3 promotes HBV replication.
FIGURE 1
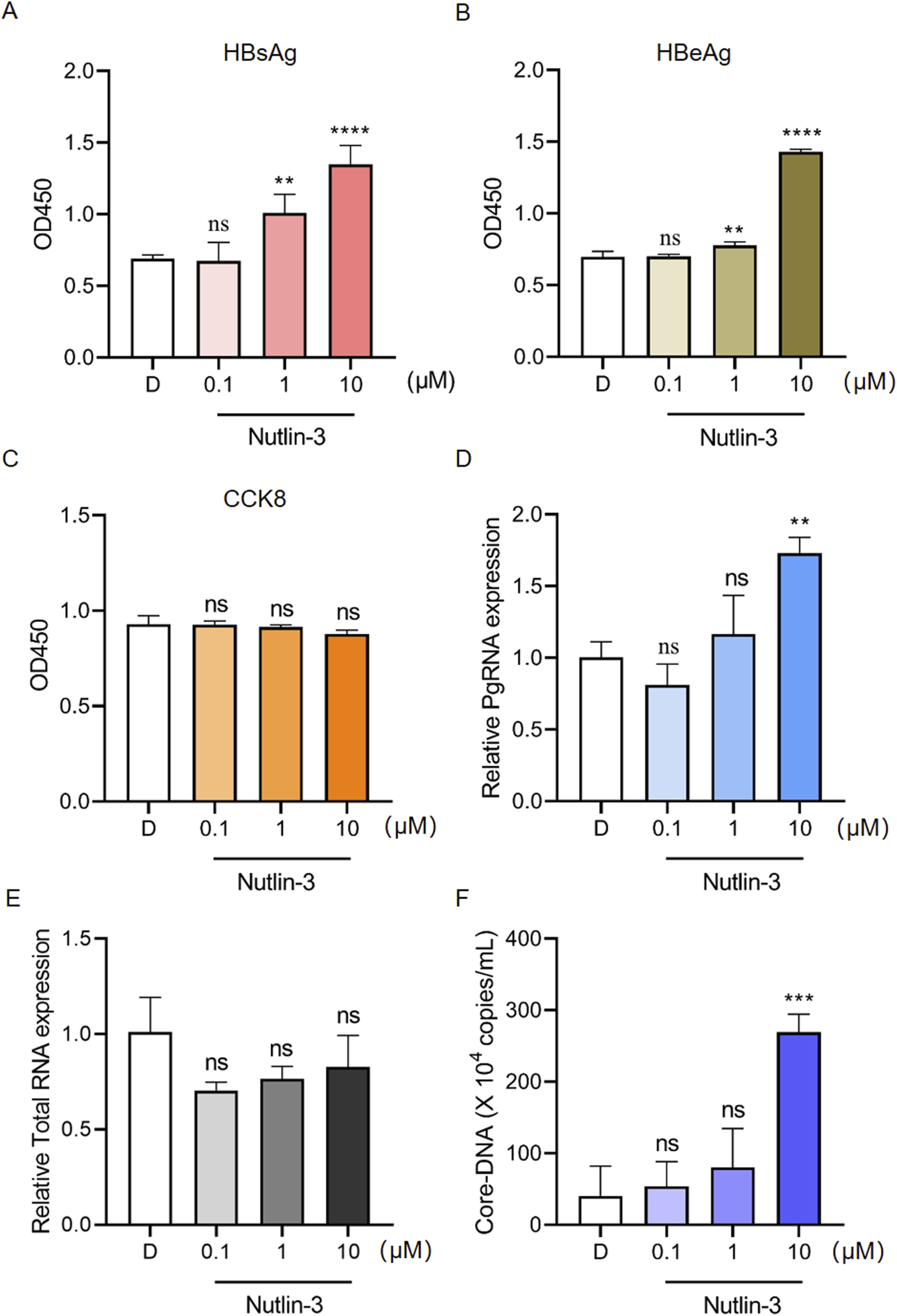
Nutlin-3 promotes HBV replication. HepG2.2.15 cells were treated with Nutlin-3 (0.1–10 μM) for 48 h. Subsequently, HBsAg (A) and HBeAg (B) levels in the culture medium were detected using ELISA. Cell viability of HepG2.2.15 cells treated with Nutlin-3 was assessed using CCK8 to evaluate the cytotoxic effects (C). HepG2.2.15 cells were treated with Nutlin-3 (0.1–10 μM) for 48 h, and then harvested for RT-qPCR analysis of HBV pgRNA (D) and total RNA (E). HepG2.2.15 cells were treated with Nutlin-3 (0.1–10 μM) for 48 h, and the core-DNA was isolated and detected using the Hepatitis B Virus Nucleic Acid Quantitative Test Kit (F). Data are presented as the mean ± standard deviation. **P < 0.01, ***P < 0.001, ****P < 0.0001, compared to the D group, where D represents DMSO treatment. ns indicates no significant effect.
The promoting effect of Nutlin-3 on HBV replication is independent of HDM2 but partially dependent on p53
To elucidate the mechanism by which Nutlin-3 promotes HBV replication, we initially examined the expression of the target proteins HDM2 and p53 by Nutlin-3. As Figures 2A–C shown, Nutlin-3 significantly increased the expression levels of HDM2 and p53 proteins. As previously mentioned, both HDM2 and p53 have the potential to influence HBV replication. We individually generated HepG2.2.15 cell lines with knockdown of HDM2 and p53, and RT-qPCR experiments confirmed the efficiency of the knockdown (Figures 2D, F). ELISA results show that knocking down HDM2 does not alter Nutlin-3’s ability to enhance HBV replication (Figure 2E). Additionally, with intact p53, Nutlin-3 increased HBsAg expression by 2.4-fold; however, following p53 knockdown, this enhancement was reduced to 1.5-fold, demonstrating that p53 knockdown partially mitigates Nutlin-3’s promotional effect on HBV (Figure 2G).
FIGURE 2
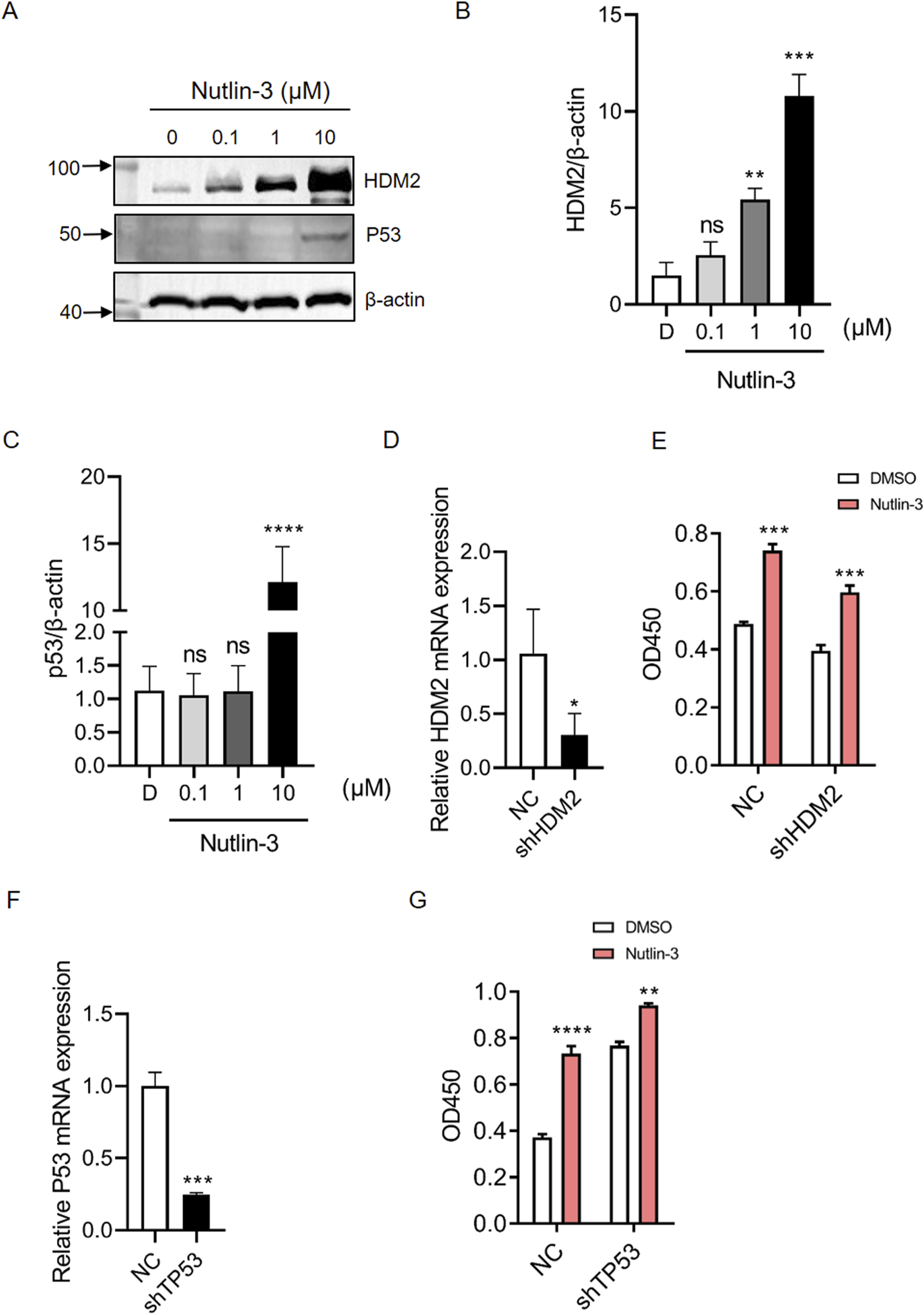
The promoting effect of Nutlin-3 on HBV replication is independent of HDM2 but partially dependent on p53. HepG2.2.15 cells were treated with Nutlin-3 (10 μM) for 48 h, protein levels of HDM2 and p53 were detected by Western blotting (A). HDM2 protein expression were analyzed with β-actin as a loading control (B). p53 protein expression were analyzed with β-actin as a loading control (C). Knockdown efficiency of HDM2 was assessed by RT-qPCR (D). HepG2.2.15 cells with HDM2 knocked-down and control cells were treated with Nutlin-3 (10 μM), with DMSO as the control. After 48 h, the levels of HBsAg in the cell supernatant were measured using ELISA (E). Knockdown efficiency of TP53 was assessed by RT-qPCR (F). HepG2.2.15 cells with TP53 knocked-down and control cells were treated with Nutlin-3 (10 μM), with DMSO as the control. After 48 h, the levels of HBsAg in the cell supernatant were measured using ELISA (G). Data are presented as the mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, compared to the D/NC group, where D represents DMSO treatment and NC represents negative control. ns indicates no significant effect.
Nutlin-3 promotes HBV replication by inhibiting the mammalian target of rapamycin (mTOR)
Several studies have shown that the activation of the mTOR pathway can significantly inhibit HBV replication (Guo et al., 2007; Teng et al., 2011). It was also reported that Nutlin-3 suppresses the mTOR pathway by promoting the expression of p53 family proteins such as p53 and p57 (Drakos et al., 2009; Shen and Maki, 2011). To determine whether Nutlin-3 promotes HBV replication by inhibiting the mTOR pathway, we used Torin 1 (a classical mTOR inhibitor) to block mTOR pathway. Without Torin 1 treatment, Nutlin-3 significantly boosted HBsAg secretion by approximately 2-fold; however, when Torin 1 was added, this effect diminishes to a mere 1.2-fold increase, which does not reach statistical significance compared to the DMSO control group (p = 0.0519). Therefore, under the influence of Torin 1, Nutlin-3 does not further enhance HBV replication (Figure 3A). The activation status of the mTOR pathway is determined by detecting the phosphorylation of its downstream proteins, S6K1 and 4E-BP1. As Figures 3B–E shown, Nutlin-3 significantly inhibited the phosphorylation of S6K1 and 4E-BP1. As a positive control, Torin 1 significantly reduced the phosphorylation of S6K1 and 4E-BP1. While Nutlin-3 did not further suppress the phosphorylation of S6K1, it somewhat enhanced the inhibitory effect on 4E-BP1 phosphorylation. This increased suppression of 4E-BP1 phosphorylation could result from a synergistic interaction between Torin 1 and Nutlin-3, likely involving overlapping signaling pathways that affect the mTOR signaling cascade, which is critical for regulating 4E-BP1 activity.
FIGURE 3
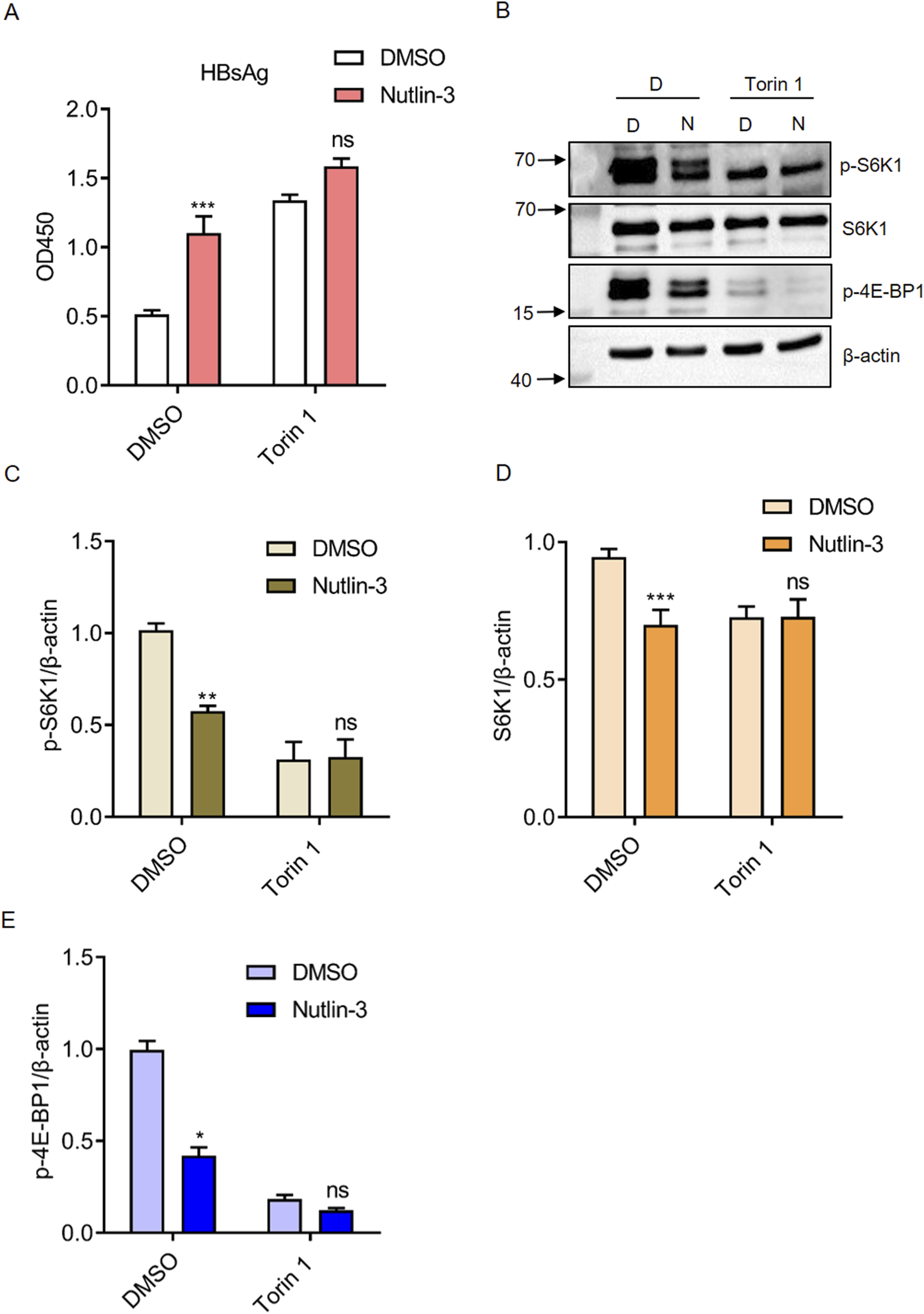
Nutlin-3 promotes HBV replication by inhibiting the mTOR pathway. HepG2.2.15 cells were treated with Nutlin-3 (10 μM) alone or in combination with Torin1 (0.1 μM) for 48 h. HBsAg levels in the culture medium were measured using ELISA (A). Protein expression of p-S6K1, S6K1, p-4E-BP1 were assessed by Western blotting (B). Relative protein levels of p-S6K1 (C), S6K1 (D), and p-4E-BP1 (E) were analyzed using ImageJ with β-actin as control. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the DMSO group. ns indicates no significant effect.
The promotion of HBV replication by Nutlin-3 depends on its activation of the autophagy pathway
Studies have indicated that autophagy plays a regulatory role in HBV replication (Tian et al., 2011), and the mTOR pathway inhibits autophagy (Kim and Guan, 2015). Therefore, we hypothesized that Nutlin-3 promotes HBV replication by inhibiting mTOR pathway, thus promoting autophagy. Autophagy is a cellular process that degrades and recycles cellular components through the formation of double-membrane vesicles called autophagosomes (Aman et al., 2021). Key markers for monitoring autophagy include LC3 and p62. LC3 (Light Chain 3) is involved in autophagosome formation, transitioning from its cytoplasmic form, LC3I, to the membrane-bound form, LC3II, which associates with autophagosome membranes. p62, also known as sequestosome 1 (SQSTM1), binds to ubiquitinated proteins, serving as a link between these proteins and the autophagy machinery, facilitating their degradation in autophagosomes. The levels of both LC3II and p62 are commonly measured to assess autophagic activity, with decreases in p62 and increases in the LC3II/LC3I ratio indicating enhanced autophagic flux. We found that Nutlin-3 significantly increased the LC3II/LC3I ratio while reducing the expression of p62 (Figures 4A–C). This suggests that Nutlin-3 promotes autophagic flux. We employed HepG2 cells stably expressing mCherry-GFP-LC3B to further investigate the activating effect of Nutlin-3 on autophagic flux. The rationale behind using mCherry-EGFP-LC3B as an indicator of autophagy is as follows (Liu et al., 2014): Under non-autophagic conditions, mCherry-EGFP-LC3B appears as diffuse yellow fluorescence in the cytoplasm, a result of the combined emission from mCherry and EGFP, when viewed under a fluorescence microscope. During autophagy, mCherry-EGFP-LC3B localizes to autophagosome membranes, forming visible yellow puncta (LC3B dots or punctae). Upon fusion of autophagosomes with lysosomes, the partial quenching of the EGFP fluorescence leads to the emergence of red dots, indicating the progression of autophagic processes. Confocal microscopy results demonstrated that Nutlin-3 significantly promoted autophagic flux (Figure 4D). Chloroquine (CQ) is a pharmacological agent that inhibits autophagy by preventing the fusion of autophagosomes with lysosomes, thereby blocking the degradation of autophagic contents (Mauthe et al., 2018). When autophagy pathway was blocked by CQ, the effect of Nutlin-3 in promoting HBsAg secretion was blocked (Figure 4E).
FIGURE 4
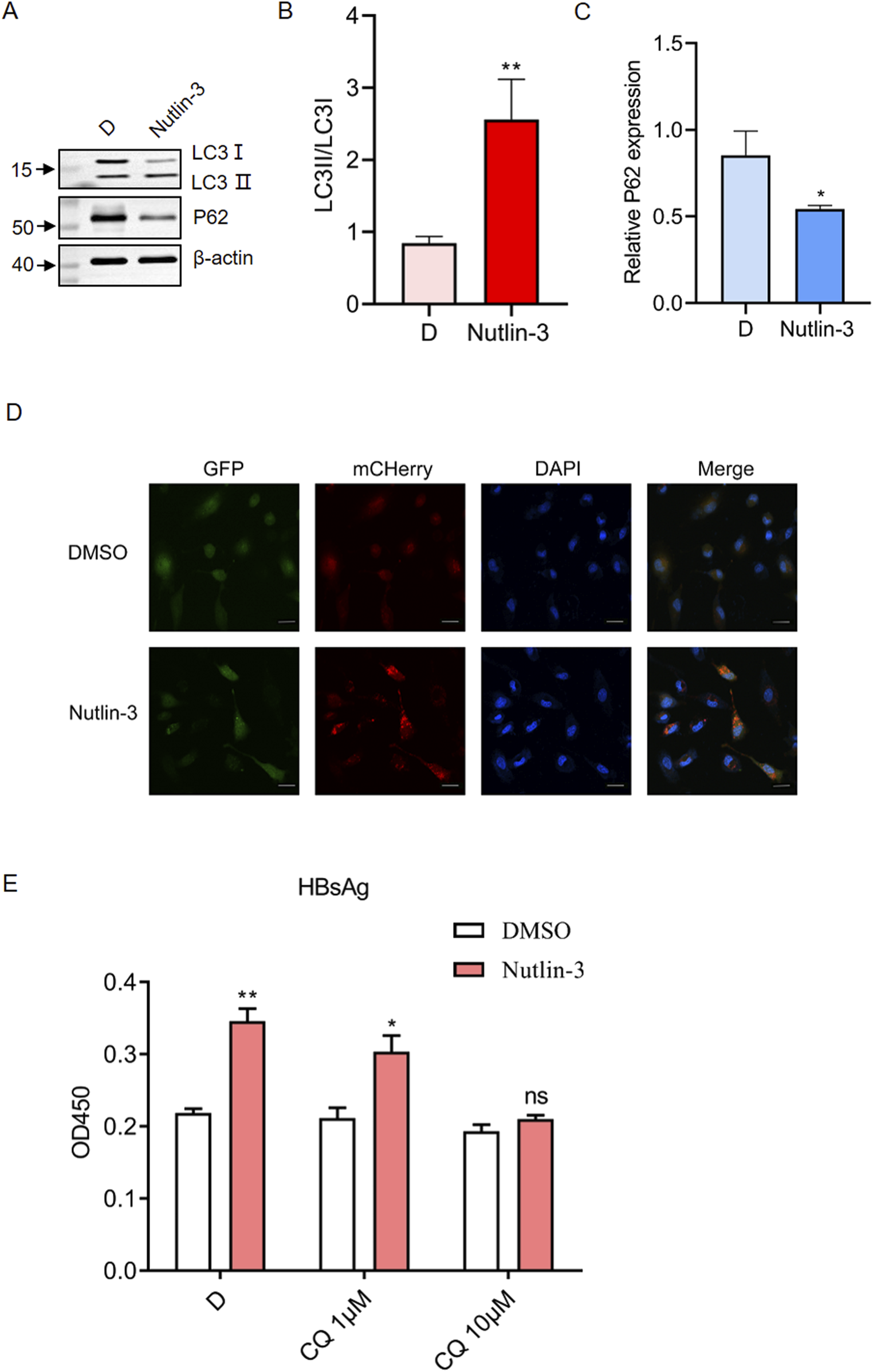
The promotion of HBV replication by Nutlin-3 depends on its activation of the autophagy pathway. HepG2.2.15 cells were treated with Nutlin-3 (10 μM) for 48 h, the protein expression of LC3B and p62 was detected by Western blotting, with β-actin as control (A). Relative protein levels of LC3Ⅱ/LC3Ⅰ (B) and p62 (C) were analyzed using ImageJ with β-actin as control. HepG2 cells stably transfected with mCherry-GFP-LC3B were treated with Nutlin-3 (10 μM) for 48 h. The expression of mCherry and GFP was assessed using confocal microscopy (D). Scale bar: 20 μm. HepG2.2.15 cells were treated with Nutlin-3 (10 μM) alone or in combination with CQ (1.10 μM) for 48 h. HBsAg levels in the culture medium were measured using ELISA (E). *P < 0.05, **P < 0.01, compared to the DMSO group. ns indicates no significant effect.
Discussion
Inhibiting the interaction between HDM2 and p53 to activate the p53 pathway represents a promising therapeutic strategy for cancer treatment. To date, numerous potent and selective small molecule inhibitors of HDM2 have been identified, with nine of them currently undergoing clinical trials, such as RG7112, Idasanutlin, SAR405838, Milademetan (Lu et al., 2023). However, these inhibitors increase the expression of HDM2 and p53 which are involved in regulating HBV replication, they therefore may influence HBV replication. As the first reported inhibitor of the HDM2 and p53 interaction, Nutlin-3 has been extensively documented in cancer research, and serving as a model molecule for studying relevant mechanisms. Therefore, we investigated the impact of Nutlin-3 on HBV replication in this study. Our data suggests that Nutlin-3 significantly promote HBV replication in HepG2.2.15 cells. Nutlin-3 significantly enhances the expression of HBsAg and HBeAg at a concentration of 10 μM. It also promotes the expression of pgRNA and Core-DNA, while having minimal impact on total RNA expression of HBV. One possible explanation for Nutlin-3 not affecting total RNA expression is that it may specifically target the transcriptional processes that boost pgRNA production without impacting the synthesis of other HBV RNAs. Further research is needed to investigate how Nutlin-3 interacts with HBV transcription factors and the specific pathways involved in pgRNA synthesis.
In the mechanistic study, we first evaluated the roles of HDM2 and p53 in the promotion of HBV replication by Nutlin-3. We confirmed that Nutlin-3 indeed promoted the expression of both HDM2 and p53 proteins through Western blotting. Through shRNA technology, we found that knocking down HDM2 did not affect the promotion of HBV by Nutlin-3. However, partial inhibition of Nutlin-3’s promotion of HBV replication was observed when p53 was knocked down. It has been reported that p53 may inhibit tumor growth through downregulating the mTOR pathway (Cui et al., 2021), and the mTOR pathway plays a crucial role in regulating HBV replication. We found that Nutlin-3 significantly inhibits the phosphorylation of S6K1 and 4E-BP1. When mTOR pathway is inhibited by small-molecule inhibitor, the promotional effect of Nutlin-3 on HBV replication is blocked. Autophagy is a conservative “self-eating” process responsible for clearing cellular components, including misfolded proteins, damaged organelles, and various invading pathogens (Parzych and Klionsky, 2014). Previous studies have shown that the replication of HBV relies significantly on the functionality of the autophagic machinery. In the present study, we confirmed that Nutlin-3 promotes autophagic flux, and when autophagy is suppressed, the promoting effect of Nutlin-3 on HBV replication is impeded.
In summary, our research has identified that Nutlin-3 can promote HBV replication by activating autophagy through the inhibition of the mTOR pathway (Figure 5). With approximately 240 million individuals worldwide still suffering from chronic HBV infection, it is crucial to be mindful of the potential risk of activating HBV replication when using small molecule drugs that inhibit the HDM2-p53 interaction in future treatments for these patients.
FIGURE 5
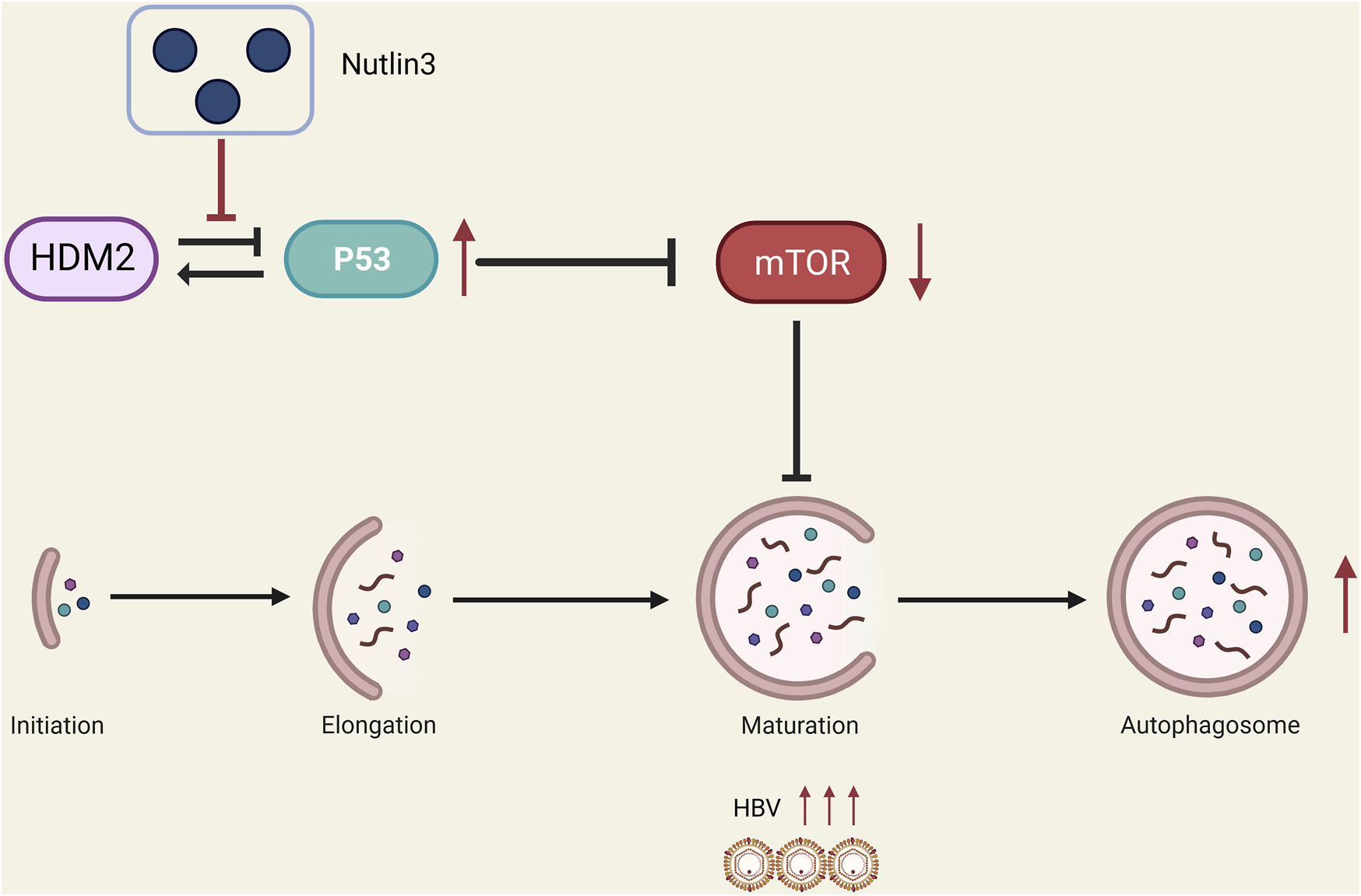
A proposed model suggests that Nutlin-3 effectively enhances HBV replication by inducing autophagic stress through the inhibition of the mTOR signaling pathway (figure created with BioRender.com).
Use of artificial intelligence tools
ChatGPT3.5 was used to improve the readability and language of the manuscript. We reviewed and edited the content as needed, taking full responsibility for the content of the publication.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
YZ: experiment design; YZ, SH, YL, and JW: acquisition and analysis of data; YZ, SH, and YL: edited the manuscript with important intellectual content; YZ and YL: obtained the funding; YZ and SH: wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Youth Program of Guangxi Natural Science Foundation of China (2021GXNSFBA196016 and 2022GXNSFBA035647), the National Natural Science Foundation of China (82360390), the Special Project Guangxi Science and Technology Base and Talent (GUIKE AD20238022), the Special Fund of the Central Government Guiding Local Scientific and Technological Development by Guangxi Science and Technology Department (GuikeZY21195024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2024.12585/full#supplementary-material
Abbreviations
HBV, Hepatitis B virus; HDM2, human double minute 2; HBsAg, Hepatitis B surface antigen; HBeAg, Hepatitis B e antigen; pgRNA, pregenomic RNA; HBx, Hepatitis X protein; pcRNA, precore RNA; mTOR, the mammalian target of rapamycin; CQ, Chloroquine.
References
1
Aman Y. Schmauck-Medina T. Hansen M. Morimoto R. I. Simon A. K. Bjedov I. et al (2021). Autophagy in healthy aging and disease. Nat. aging1, 634–650. 10.1038/s43587-021-00098-4
2
Block T. M. Zhou T. Anbarasan N. Gish R. (2016). Evolving new strategies for the medical management of chronic hepatitis B virus infection. Gastroenterol. Hepatol. (N Y)12, 679–689.
3
Cui D. Qu R. Liu D. Xiong X. Liang T. Zhao Y. (2021). The cross talk between p53 and mTOR pathways in response to physiological and genotoxic stresses. Front. cell Dev. Biol.9, 775507. 10.3389/fcell.2021.775507
4
Dey A. Wong E. T. Bist P. Tergaonkar V. Lane D. P. (2007). Nutlin-3 inhibits the NFκB pathway in a p53 dependent manner: implications in lung cancer therapy. Cell Cycle6, 2178–2185. 10.4161/cc.6.17.4643
5
Drakos E. Atsaves V. Li J. Leventaki V. Andreeff M. Medeiros L. J. et al (2009). Stabilization and activation of p53 downregulates mTOR signaling through AMPK in mantle cell lymphoma. Leukemia23, 784–790. 10.1038/leu.2008.348
6
Fung S. Choi H. S. J. Gehring A. Janssen H. L. A. (2022). Getting to HBV cure: the promising paths forward. Hepatology76, 233–250. 10.1002/hep.32314
7
Guo H. Zhou T. Jiang D. Cuconati A. Xiao G. H. Block T. M. et al (2007). Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J. Virol.81, 10072–10080. 10.1128/jvi.00541-07
8
Haupt Y. Maya R. Kazaz A. Oren M. (1997). Mdm2 promotes the rapid degradation of p53. Nature387, 296–299. 10.1038/387296a0
9
Kim Y. C. Guan K. L. (2015). mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest.125, 25–32. 10.1172/jci73939
10
Levine A. J. (1997). p53, the cellular gatekeeper for growth and division. Cell88, 323–331. 10.1016/s0092-8674(00)81871-1
11
Liu B. Liu C. Zhao X. Shen W. Qian L. Wei Y. et al (2014). Establishment of a cell line with stable expression of mCherry-EGFP tandem fluorescent-tagged LC3B for studying the impact of HIV-1 infection on autophagic flux. J. virological methods209, 95–102. 10.1016/j.jviromet.2014.09.012
12
Liu N. Zhang J. Yang X. Jiao T. Zhao X. Li W. et al (2017). HDM2 promotes NEDDylation of hepatitis B virus HBx to enhance its stability and function. J. Virol.91, 003400-17. 10.1128/jvi.00340-17
13
Lu Y. Wu M. Xu Y. Yu L. (2023). The development of p53-targeted therapies for human cancers. Cancers15, 3560. 10.3390/cancers15143560
14
Ma C. Xu W. Yang Q. Liu W. Xiang Q. Chen J. et al (2020). Osteopetrosis-associated transmembrane protein 1 recruits RNA exosome to restrict hepatitis B virus replication. J. Virol.94, e01800-e01819. 10.1128/jvi.01800-19
15
Mauthe M. Orhon I. Rocchi C. Zhou X. Luhr M. Hijlkema K. J. et al (2018). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy14, 1435–1455. 10.1080/15548627.2018.1474314
16
Ori A. Zauberman A. Doitsh G. Paran N. Oren M. Shaul Y. (1998). p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J.17, 544–553. 10.1093/emboj/17.2.544
17
Parzych K. R. Klionsky D. J. (2014). An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal20, 460–473. 10.1089/ars.2013.5371
18
Psatha K. Kollipara L. Drakos E. Deligianni E. Brintakis K. Patsouris E. et al (2023). Interruption of p53-MDM2 interaction by nutlin-3a in human lymphoma cell models initiates a cell-dependent global effect on transcriptome and proteome level. Cancers15, 3903. 10.3390/cancers15153903
19
Sells M. A. Chen M. L. Acs G. (1987). Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. U. S. A.84, 1005–1009. 10.1073/pnas.84.4.1005
20
Shen H. Maki C. G. (2011). Pharmacologic activation of p53 by small-molecule MDM2 antagonists. Curr. Pharm. Des.17, 560–568. 10.2174/138161211795222603
21
Supiot S. Hill R. P. Bristow R. G. (2008). Nutlin-3 radiosensitizes hypoxic prostate cancer cells independent of p53. Mol. cancer Ther.7, 993–999. 10.1158/1535-7163.mct-07-0442
22
Tabe Y. Sebasigari D. Jin L. Rudelius M. Davies-Hill T. Miyake K. et al (2009). MDM2 antagonist nutlin-3 displays antiproliferative and proapoptotic activity in mantle cell lymphoma. Clin. Cancer Res.15, 933–942. 10.1158/1078-0432.ccr-08-0399
23
Teng C. F. Wu H. C. Tsai H. W. Shiah H. S. Huang W. Su I. J. (2011). Novel feedback inhibition of surface antigen synthesis by mammalian target of rapamycin (mTOR) signal and its implication for hepatitis B virus tumorigenesis and therapy. Hepatology54, 1199–1207. 10.1002/hep.24529
24
Tian Y. Sir D. Kuo C. F. Ann D. K. Ou J. H. (2011). Autophagy required for hepatitis B virus replication in transgenic mice. J. Virol.85, 13453–13456. 10.1128/jvi.06064-11
25
Vassilev L. T. Vu B. T. Graves B. Carvajal D. Podlaski F. Filipovic Z. et al (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science303, 844–848. 10.1126/science.1092472
26
Vogelstein B. Lane D. Levine A. J. (2000). Surfing the p53 network. Nature408, 307–310. 10.1038/35042675
27
Voon Y. L. Ahmad M. Wong P. F. Husaini R. Ng W. T. Leong C. O. et al (2015). Nutlin-3 sensitizes nasopharyngeal carcinoma cells to cisplatin-induced cytotoxicity. Oncol. Rep.34, 1692–1700. 10.3892/or.2015.4177
28
Yardeni D. Chang K. M. Ghany M. G. (2023). Current best practice in hepatitis B management and understanding long-term prospects for cure. Gastroenterology164, 42–60.e6. 10.1053/j.gastro.2022.10.008
29
Yee-Lin V. Pooi-Fong W. Soo-Beng A. K. (2018). Nutlin-3, A p53-mdm2 antagonist for nasopharyngeal carcinoma treatment. Mini Rev. Med. Chem.18, 173–183. 10.2174/1389557517666170717125821
30
Zheng T. Yin D. Lu Z. Wang J. Li Y. Chen X. et al (2014). Nutlin-3 overcomes arsenic trioxide resistance and tumor metastasis mediated by mutant p53 in Hepatocellular Carcinoma. Mol. Cancer13, 133. 10.1186/1476-4598-13-133
Summary
Keywords
Nutlin-3, HBV, mTOR, autophagy, p53
Citation
Han S, Li Y, Wang J and Zhang Y (2024) Nutlin-3 promotes HBV replication by activating mTOR-mediated autophagy. Acta Virol. 68:12585. doi: 10.3389/av.2024.12585
Received
19 December 2023
Accepted
14 November 2024
Published
22 November 2024
Volume
68 - 2024
Edited by
Ivana Nemčovičová, Slovak Academy of Sciences, Slovakia
Reviewed by
Pavlina Bartikova, Slovak Academy of Sciences, Slovakia
Vladimir Zelnik, Slovak Academy of Sciences, Slovakia
Updates
Copyright
© 2024 Han, Li, Wang and Zhang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhang, zhangyong@glmc.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.