Abstract
Persistent infection with gammaherpesviruses, such as Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus may lead to development of various lymphomas and carcinomas in humans. Murid gammaherpesvirus 4 (MuHV-4, also known as MHV-68) is widely used as a model to study all aspects of gammaherpesvirus pathogenesis. Similar to human gammaherpesviruses, MuHV-4 is known for its variability, as evidenced by the existence of multiple strains/variants that differ from each other in primary sequence and some pathogenetic features. Detailed knowledge of the dependence of pathogenic properties on genome structure and sequence would help to better understand the complex host-pathogen interactions. The main aim of this study was to analyze how various strains/variants of MuHV-4, mainly MHV-72 and MHV-4556 induce latency and reactivation. In this study, we characterized the ability of MHV-72 and MHV-4556 to establish latency in spleen, lungs and thymus of immunocompetent mice after intranasal infection. Moreover, we described the potential of these strains to reactivate ex vivo using the method of tissue explantation and stimulation with inhibitor of histone deacetylases, trichostatin A. Our results clearly demonstrate that both MHV-72 and MHV-4556 exhibit a significant deficit in the establishment of deep latency in spleen. Moreover, MHV-72 has a reduced ability to establish latency in lungs as well as thymus. Regarding reactivation, we found that both MHV-72 and MHV-4556 have a significant deficit in ex vivo reactivation from latency in lungs as well as thymus in comparison to MHV-68. Moreover, MHV-72 also possessed a deficit in the ex vivo reactivation from spleen. In contrast, MHV-4556 was able to respond to the trichostatin A-mediated reactivation stimuli by increasing the DNA copy number but had a defect in the release from tissue during the early and deep latency in spleen. Our results contribute to molecular characterization of differences among the MuHV-4 strains in the ability to establish latency and reactivation and could also serve to better understanding of strain-dependent variances among human gammaherpesviruses.
Introduction
Two human gammaherpesviruses, Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), are classified as class I carcinogens by the International Agency for Research on Cancer because of their causal links to carcinomas and malignant lymphoproliferative diseases. Worldwide, EBV persists in more than 95% of the adult human population. The seroprevalence of KSHV is more than 50% in sub-Saharan Africa, and less than 10% in northern Europe and North America (de Martel et al., 2020).
To date, several strains of EBV and KSHV have been isolated in all parts of the world, differing not only in genomic sequence but also in pathogenesis and ability to contribute to tumorigenesis. Analysis of EBV isolates from nasopharyngeal carcinoma biopsies using next-generation sequencing revealed that although all EBV isolates are very similar to the prototypic EBV, there are differences in several genes that may lead to phenotypic and functional differences (Su et al., 2023). In the case of KSHV, which has so far been subtyped mainly on the basis of sequence differences in the ORF-K1, ORF-K18 and ORF-73 genes, whole genome sequencing within the more conserved region of the KSHV genome has identified four additional genes containing polymorphisms that contribute significantly to differences in gene function and pathogenesis between KSHV isolates (Olp et al., 2015; Jary et al., 2021).
Murine gammaherpesvirus 68 (MHV-68), a natural pathogen of murid rodents belonging to the species Murid gammaherpesvirus 4 (MuHV-4), is classified in the genus Rhadinovirus, the subfamily Gammaherpesvirinae, and is genetically closely related to human oncogenic gammaherpesviruses EBV and KSHV. Because the pathogenesis of MHV-68 is similar to that of EBV and KSHV, MHV-68 has been widely used as a model to study many aspects of human gammaherpesvirus pathogenesis, including lymphoproliferation and tumor development. In addition to MHV-68, seven murine gammaherpesviruses designated −60, −72, −76, −78, −4556, −5682, and -ŠUM were isolated from wild-living Clethrionomys and Apodemus rodents in Slovakia and thereafter in the Czech Republic (Blaskovic et al., 1980; Blaskovic and Mistrikova, 1985; Kozuch et al., 1993). Generally, all isolates are very similar, nevertheless, they differ from each other in some pathogenic properties related to differences in primary sequence of several genes as well as in a variability of the left end of their genomes (Halásová et al., 2011; Kúdelová et al., 2012; Čipková-Jarčušková et al., 2013; Mistríková and Briestenská, 2020). All these differences could lead to distinct functionality of corresponding proteins, and thus account to distinct host-pathogen interactions. It was shown that all strains/isolates of MuHV-4, including the prototype MHV-68, possess very similar pathological features of the acute phase of infection (Macrae et al., 2001; Čipková-Jarčušková et al., 2013). Therefore, we suggest that the key differences among the strains of MuHV-4 should be seen not during the productive infection, but in latency and reactivation.
The main aim of this study was to characterize the strain-dependent ability of MuHV-4 to induce latency and reactivate from latency. We evaluated the ability of MHV-72 (clone h3.7) and MHV-4556 (clone i2.8) to establish latency in lungs, spleen, and thymus of immunocompetent mice after intranasal infection. The time course of the onset of latency we studied from day 10, which represents the peak of the acute phase of infection, through days 14 and 21, which can be considered the early latency stage, to days 35 and 42, when the deep latency is established. Our results clearly demonstrate that both, MHV-72 and MHV-4556, exhibit a significant deficit in the establishment of deep latency in spleen. Moreover, MHV-72 has a reduced ability to latently infect the lungs as well as thymus. To characterize the ability of MHV-72 and MHV-4556 to reactivate from latency, we used the ex vivo tissue explantation method (Rajcani et al., 1985) in the presence of trichostatin A (TSA), which is able to induce passive demethylation of the Rta promoter of MHV-68 (Tsuji et al., 1976; Yang et al., 2009; Lapuníková et al., 2015). We found that both, MHV-72 and MHV-4556, have a significant deficit in ex vivo reactivation from latency in lungs as well as thymus in comparison to MHV-68. Moreover, MHV-72 also possessed a deficit in the ex vivo reactivation from spleen. In contrast, MHV-4556 was able to respond to the reactivation stimuli, which was observed as increased DNA copy number but had a defect in the release from tissue during the early and deep latency in spleen. Our results contribute to the existing knowledge on the pathogenesis of individual MuHV-4 strains/isolates by molecular characterization of the differences between MHV-68, MHV-72, and MHV-4556 in their ability to establish latency and to reactivate from latency ex vivo.
Materials and methods
Cell lines and viruses
The Vero (ATCC® CCL-81™) and BHK-21[C-13] (ATCC® CCL-10TM) cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, United States) supplemented with 5% (v/v) fetal bovine serum (FBS) (HyClone, United States), 2 mmol/l L-glutamine, 100 U of streptomycin per ml and 100 U of penicillin per ml (Lonza, Belgium) and incubated at 37°C in 5% CO2 atmosphere. Experiments were performed with three viruses: MHV-68 clone f2.6, MHV-72 clone h3.7 - GeneBank Acc. No.: MW256631.1 (Raslova et al., 2000), and MHV-4556 clone i2.8- GeneBank Acc. No.: MW256632.1 (Valovičová et al., 2006). Working virus stocks were prepared by infection of BHK-21 cells at a multiplicity of infection (MOI) of 0.05 PFU/cell. The virus stocks were stored at −80°C until further use. All experiments with virus were performed at biosafety level 2.
Mice
Six weeks old BALB/c mice were purchased from Laboratory Animal Breeding and Experimental Facility of Faculty of Medicine, Masaryk University in Brno, Czech Republic. Mice were bred and housed in specific pathogen-free barriers facility in accordance with institutional and state guidelines. All animal experiments were approved by The State Veterinary and Food Administration of the Slovak Republic under permit number Ro. 3497/14-221.
Intranasal infection of mice
Mice were divided into 4 groups (MHV-68 infected; MHV-72 infected; MHV-4556 infected; uninfected), with each group consisting of 25 mice. Mice were infected by intranasal administration of 2 × 105 PFU virus in 25 μL DMEM during light isoflurane anesthesia. At 10, 14, 21, 35, and 42 days post infection (dpi), mice were sacrificed by cervical spine dislocation under deep isoflurane anesthesia. Five mice from each group were tested at each time interval. The spleen, thymus, and lung were harvested, weighed, and each organ was divided into two equal parts. One part of the organs was immediately used for single cell suspension (SCS) preparation followed by infectious center assay (ICA) and viral genome copy number determination by qPCR, and the other part was used for the ex vivo tissue explantation method in the presence of TSA.
Ex vivo tissue explantation and viral reactivation induced by trichostatin A
One half of each organ (spleen, thymus, lung) was sterilely sectioned into several small pieces (size approximately 2–3 mm3) and the tissue pieces were transferred to wells on a 6-well plate. The Roswell Park Memorial Institute-1640 medium (RPMI) (2 mL; Lonza), supplemented with 10% FBS, 2 mmol/l L-glutamine, 100 U of streptomycin per ml, 100 U of penicillin per ml (Lonza) and TSA (200 ng/mL; Invivogen, United States), was added and the organs were cultivated for 10 days at 37°C in 5% CO2. At 3, 7, and 10 days after explantation (dpe), the medium was removed and fresh medium of the same composition was added. The titer of virus released into the explantation medium was determined by plaque assay on Vero cells (see below). At the end of explantation (at 10 dpe), tissue pieces were collected and homogenized using a 6.35 mm diameter Zirconium Oxide-coated ceramic bead in a FastPrep-24™ High-Speed benchtop homogenizer (20 m/s/5s) (MP Biomedicals, United States). DNA was extracted from the homogenate using the QIAamp DNA mini kit (Qiagen, Germany) according to the manufacturer’s instructions. DNA was stored at −20°C until further use (see qPCR).
Plaque assay
To determine virus titer in media collected at 3, 7, and 10 dpe, Vero cells were seeded in 24-well plates (1 × 105 cells/well) and cultured in DMEM supplemented with 5% FBS at 37°C and 5% CO2 overnight. The next day, Vero cells were incubated with 100 μL of serially diluted samples for 45 min, followed by the addition of 1 mL of DMEM containing 0.43% methylcellulose. The plates were further incubated for 7–10 days at 37°C in 5% CO2. Cells were then stained with 0.03% methylene blue to quantify plaques. All samples were tested in duplicate.
Single-cell suspension and infectious center assay
One half of each organ (spleen, thymus, lung) was homogenized using a 6.35 mm diameter Zirconium Oxide-coated ceramic bead in a FastPrep-24™ High-Speed benchtop homogenizer (20 m/s/5 s) (MP Biomedicals, United States) to prepare SCS. Cells were counted and 100 μL of serially diluted suspension (5 × 105–5 cells/mL) was added to Vero cells seeded into wells (1 × 105 cells/well) 1 day prior to the assay. Cells were incubated for 14 days at 37°C in a 5% CO2 atmosphere, then fixed, stained with toluidine blue and the number of infection centers was counted. All samples were tested in duplicate. The remaining SCS was used for DNA isolation using the QIAamp DNA mini kit (Qiagen) according to the manufacturer’s instructions. DNA was stored at −20°C until further use (see qPCR).
Quantitative real-time PCR (qPCR)
Genome copy number of MHV-68, MHV-72, and MHV-4556 was quantified using the Luminaris HiGreen qPCR master mix, high ROX (ThermoScientific) according to the manufacturer’s instructions. The amplification of a 66 bp long fragment of ORF65 (M9) gene (Acc. No. NC_001826.2), a small (basic) capsid component, was performed with a pair of primers F: 5′-GTC AGG GCC CAG TCC GTA-3′ and R: 5′-TGG CCC TCT ACC TTC TGT TGA-3′ (Guo et al., 2009). The qPCR mixture contained 0.6 μmol/L of each primer and 200 ng DNA in the final volume 20 µL. The qPCR was carried out in an Applied Biosystems StepOne™ Real-Time PCR systems thermocycler (Applied Biosystems) under the following conditions: 95°C/10 min and 40x (95°C/15 s, 58°C/15 s, 72°C/15 s). All samples were tested in triplicates and at the same time with the template-free qPCR mixture (negative control). The standard curve was prepared by serial dilution of MHV-68 baculovirus DNA (Adler et al., 2000).
Statistical analysis
The data represent two independent experiments. Data are presented as a mean of log10 ± standard error of the mean (SEM). Comparison between uninfected and infected groups and between the MHV-68-infected group versus the MHV-72 and MHV-4556-infected groups were performed using a two-tailed unpaired Student’s t-test. Statistical significance is reported as either *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Results
Latency
MHV-68 enters new hosts by infection of epithelial cells. After the primary replication in epithelial cells of lungs or nasal cavity (by intranasal infection), virions preferentially infect B cells through epithelial-myeloid-lymphoid transfer. Subsequently, B-cells mediate transport of MHV-68 into internal organs, such as lung, spleen, nose, salivary glands, thymus and others. All of these infection events could serve as independent source of viral persistence and are affected by host immunity and virus pathogenicity (Frederico et al., 2012; Wang et al., 2021). This led us to investigate the organ-specific settling time and the ability of our murine gammaherpesvirus strains/isolates, MHV-72 and MHV-4556, to establish latency in the major MHV-68 persistence targets - the lungs, spleen, and thymus.
MHV-72 possesses a reduced ability to establish latency in lungs, spleen and thymus. MHV-4556 possesses a reduced ability to establish latency only in spleen
To characterize the ability of MHV-72 and MHV-4556 to induce latency in the lung, spleen, and thymus after intranasal infection in mice and compare it with MHV-68, we used two methods: ICA to detect infectious virus and qPCR to quantify copies of the viral genome in tissue (Figure 1). The time course of the onset of latency was studied from day 10, which represents the peak of the acute phase of infection, through days 14 and 21, which can be considered the early latency stage, to days 35 and 42, when the deep latency is established.
FIGURE 1
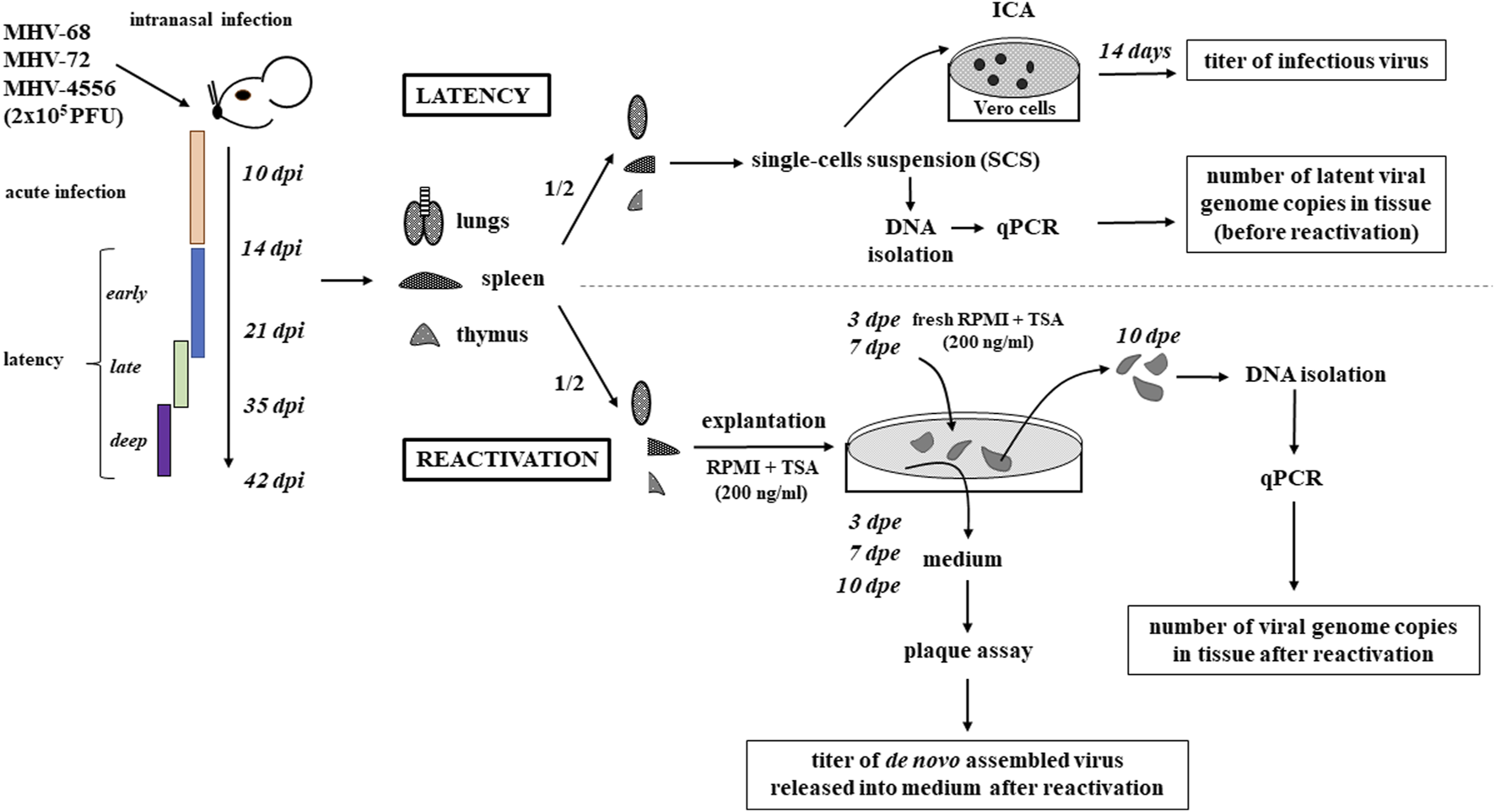
Schematic diagram of experimental design. BALB/c mice were infected intranasally with MHV-68, MHV-72 or MHV-4556 at a dosage 2 × 105 PFU. The mice were euthanized at 10, 14, 21, 35, and 42 dpi, and the lungs, spleen and thymus were collected, weighed and divided in half. One half of each organ was used for study of latency establishment; the second half was used for study of capacity for ex vivo reactivation. To study the latency establishment (time course of early, late and deep latency is shown in scheme), the single-cells suspension (SCS) was prepared by organ homogenization. The cells were counted and 100 μL of serially diluted SCS (5 × 105–5 cells/mL) was added to Vero cells. After 14 days, the number of infectious centers was counted. The rest of the SCS was used for DNA isolation and quantification of viral genome copies by qPCR. To study the capacity for ex vivo reactivation, one half of organ was explanted in RPMI medium supplemented with TSA (200 ng/mL) for 10 days. On the day 3 and 7 post explantation (dpe), the medium was removed and the fresh medium of the same composition was added. The titer of virus released into the explantation medium was determined by plaque assay not only at 3 and 7, but also at 10 dpe. At the end of the explantation (at 10 dpe), the tissues were homogenized, the DNA was extracted and the number of viral genome copies in tissue was determined by qPCR.
Using the ICA method, we found no significant differences in the amount of infectious virus present in the lungs, spleen, and thymus of mice infected with MHV-68, MHV-72, or MHV-4556. The amount of infectious virus MHV-68, MHV-72, or MHV-4556 reached a maximum of 100 PFU in the lungs at 10 dpi. Later, from 14 to 42 dpi, infectious virus was almost undetectable (max. 20 PFU/tissue) in all organs tested.
On the other hand, the quantification of viral genome copies by qPCR revealed the significant differences in the ability of MHV-72 and MHV-4556 to latently colonize the lungs, spleen and thymus (Figure 2). In comparison to MHV-68, MHV-72 had significant deficit in establishment of latency in all tested organs. The number of MHV-72 genome copies in the lungs was significantly reduced at all time points except 21 dpi, by approximately 1–2 log10 (P ≤ 0.001) (Figure 2A). In the spleen, the MHV-72 genome copy number at the end of acute infection (10 dpi) (Figure 2B) was approximately 1.5 log10 (P ≤ 0.001) higher compared with MHV-68. However, at 14 dpi and later on, we detected a significant deficit of MHV-72 in the establishment of latency in spleen. In addition to lungs and spleen, we observed reduced ability of MHV-72 to establish latency at 14 and 35 dpi also in thymus (Figure 2C).
FIGURE 2
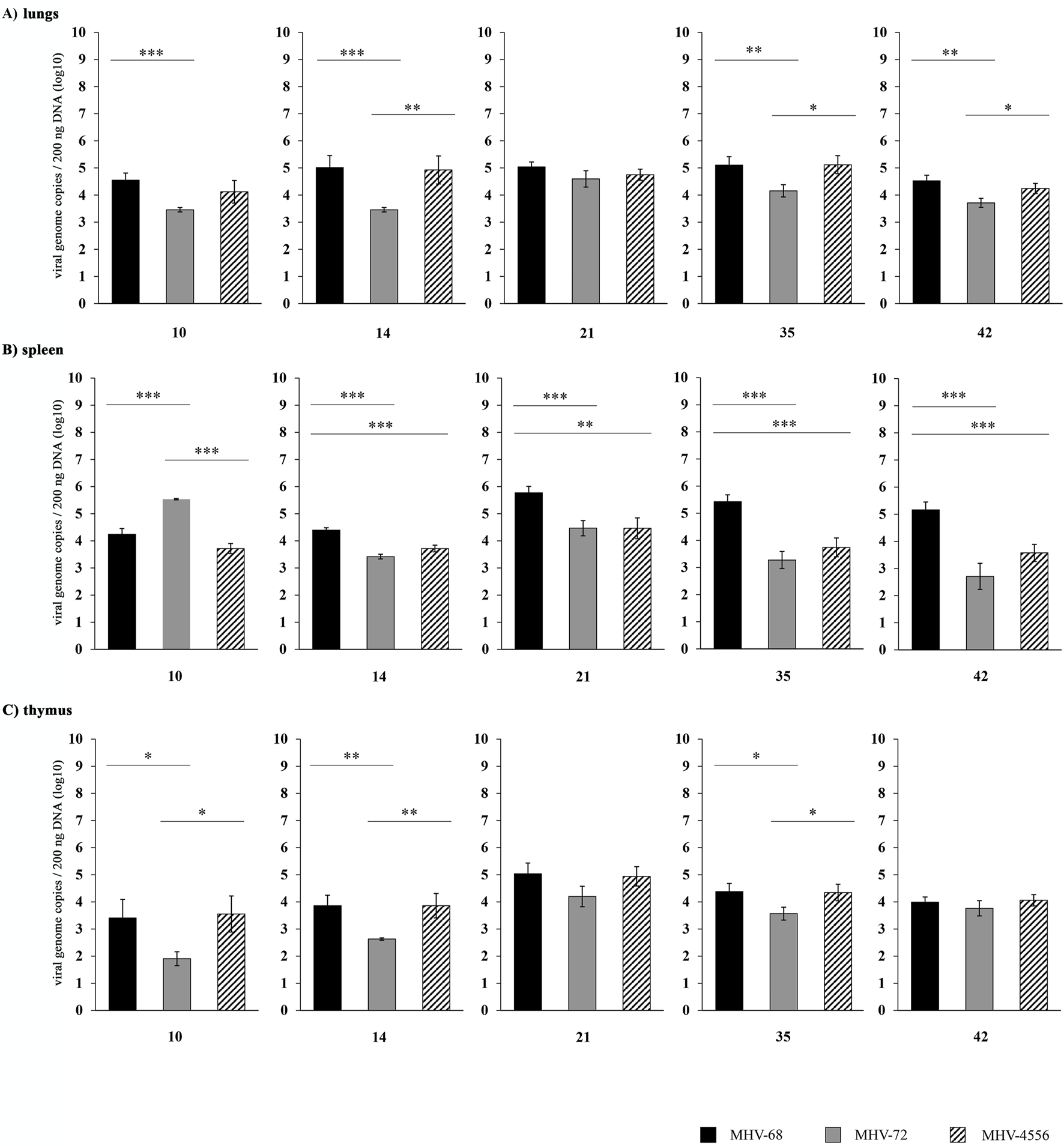
Time course of MHV-72 and MHV-4556 latency establishment in lungs, spleen and thymus BALB/c mice were infected intranasally with 2 × 105 PFU of MHV-68, MHV-72 and MHV-4556. The mice were euthanized at 10, 14, 21, 35, and 42 dpi, and the lungs, spleen and thymus were collected, weighed and divided in half. One half of each organ was used for DNA isolation and quantification of viral genome copies by qPCR. (A) Viral genome copies of MHV-68 vs. MHV-72 vs. MHV-4556 in lungs. (B) Viral genome copies of MHV-68 vs. MHV-72 vs. MHV-4556 in spleen. (C) Viral genome copies of MHV-68 vs. MHV-72 vs. MHV-4556 in thymus. Five mice per group were assayed at each time interval. Data are presented as the mean of genome copy number (log10) ± SEM and the graphs represent the average of data of two independent experiments. *P ≤ 0.05, **P ≤ 0.01 ***P ≤ 0.001.
The ability of MHV-4556 to establish latency in the lungs and thymus was similar to that of MHV-68 (Figures 2A, C). However, in the spleen, the MHV-4556 genome copy number reached a lower level compared with MHV-68 (a decrease of approximately 1–1.5 log10 (P ≤ 0.001 and P ≤ 0.01, respectively)) at 14, 21, 35, and 42 dpi (Figure 2B).
We have shown that the ability of both MHV-72 and MHV-4556 to establish latency in the spleen is lower compared to MHV-68. In addition, MHV-72 also has a lower ability to establish latency in the lungs and thymus, which distinguishes it from MHV-4556.
Reactivation
In general, latently infected cells contain a viral genome that is in a covalently closed circular (episomal) conformation and is difficult to reconstitute into an infectious form without stimulation. External stimuli, such as stress and immunodeficiency, may lead to reactivation of the virus from latency and subsequent productive infection of the host. Viral reactivation is a complex process regulated simultaneously by viral and cellular factors, including the epigenetic modifications, which have been intensively studied in recent years (Buschle and Hammerschmidt, 2020; Nehme et al., 2020; Shareena and Kumar, 2023).
Spontaneous reactivation of MHV-68 can be stimulated through the explantation of latently infected cells/tissue and their co-cultivation with permissive cells or induction of immunosuppression by removal of T cells, including CD4+ and CD8+ T cells individually or simultaneously, immunosuppression with cyclosporin A, and treatment with dexamethasone. However, only approximately 10% of latent virus is able to reactivate in these conditions (Wang et al., 2021). It was shown (Yang et al., 2009; Lapuníková et al., 2015) that TSA contributes to the start of viral gene transcription and MHV-68 reactivation through inhibition of histone deacetylases, which results in acetylation of H3 and H4 histones.
Using the ICA method, by which we could theoretically detect both infectious virus and virus in latency (Weck et al., 1996), we found that the ability of MHV-68, MHV-72 and MHV-4556 to reactivate ex vivo from SCS was very limited even after TSA treatment. Reactivation of MHV-68 was detected only from SCS of lungs explanted at day 42 p.i. and from SCS of spleen explanted at days 10, 21 and 42 p.i. For MHV-4556, we observed only low levels of reactivation from SCS of lungs explanted at day 10 p.i. and no reactivation of MHV-72 and MHV-4556 from SCS of spleen and thymus.
Rajcani et al. (1985) successfully used the explantation technique for recovery of the latent virus from different organs of mice infected with MHV-68. Therefore, we utilized this explantation technique in combination with TSA stimulation to examine the ability of the MHV-68, MHV-72, and MHV-4556 to reactivate ex vivo. Tissues of lungs, spleen and thymus were explanted in the presence of TSA (200 ng/mL) for 10 days. The number of de novo assembled infectious virus released into the explantation medium was evaluated by plaque assay at 3, 7, and 10 dpe. To detect the amount of virus not released from the explanted tissue, we quantified the number of genome copies in tissues by qPCR at 10 dpe (Figure 1). In our study, we identified that the ability of all three strains of MuHV-4 for TSA-induced reactivation from tissue explants is much higher compared to the reactivation from SCS. This suggests that intercellular communication within the tissue is required for efficient MuHV-4 reactivation, but cannot be maintained in a SCS.
Reactivation from the lungs
We found very low levels of infectious virus in the media of explanted lungs sections stimulated with TSA for 3 days, indirectly confirming our results when infectious virus was almost absent in the tissue before the reactivation procedure (see Results, part 1. Latency). Despite it, we identified that prototype MHV-68 has high ability to reactivate from lungs sections cultivated with TSA (Figure 3A). De novo assembled infectious MHV-68 reached titer up to 4 log10 PFU/tissue at 7 dpe and 6 log10 PFU/tissue at 10 dpe in tissue collected 14 and 21 dpi. High ability of MHV-68 to reactivate was also confirmed by qPCR (Figure 3B), when we detected approximately two-times higher number of genome copies after the TSA-induced reactivation in comparison to the level of genome copies in latency (before TSA stimulation). However, the level of MHV-68 reactivation was continuously decreasing with the progression of latency. During the deep latency (at 35 and 42 dpi), we detected significant reduction in titer of de novo assembled MHV-68 (<4 log10 PFU/tissue) as well as lower numbers of genome copies (<10 log10 viral genome copies/200 ng DNA) in lungs sections (Figures 3A, B).
FIGURE 3
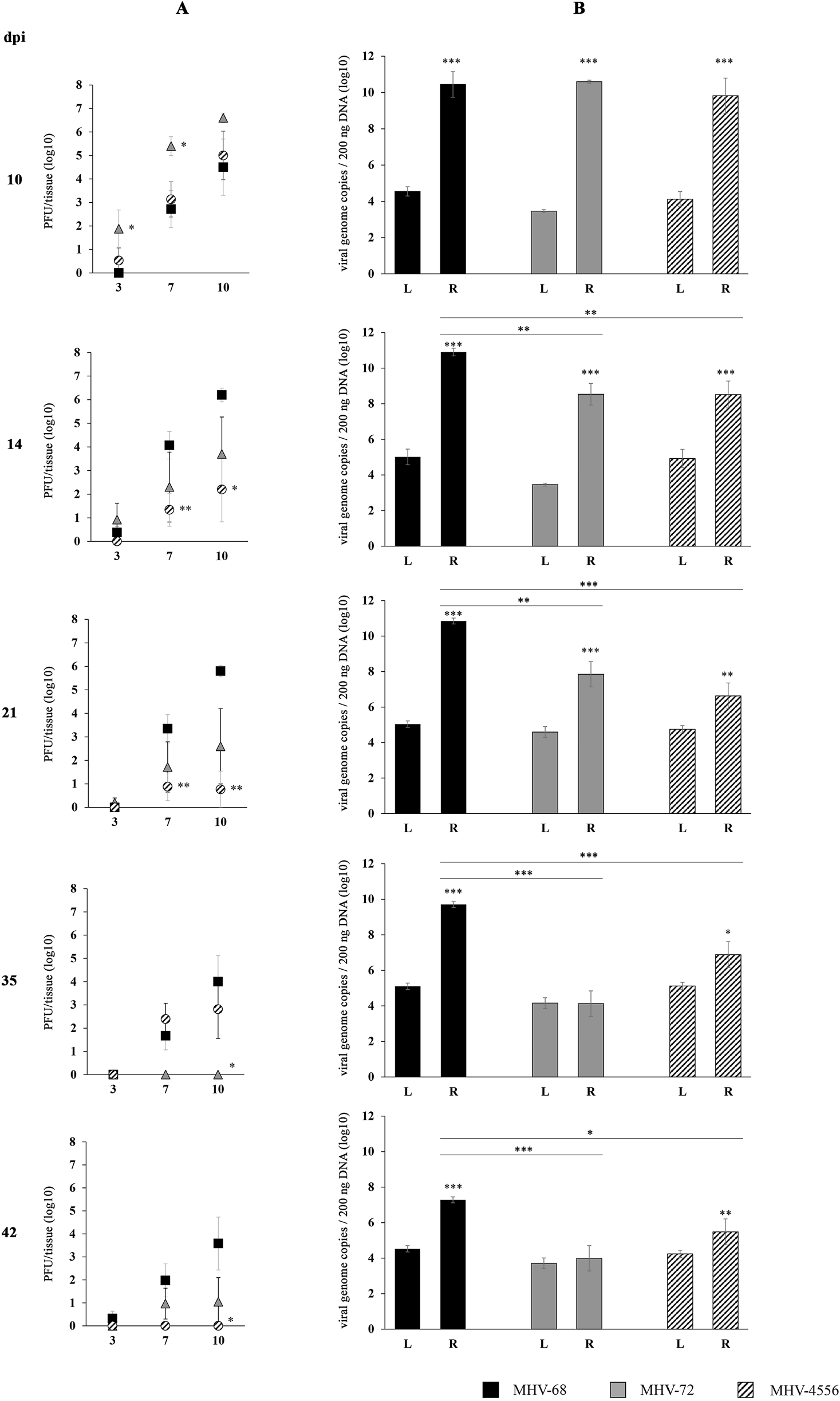
TSA-induced ex vivo reactivation of MHV-72 and MHV-4556 in the lungs. BALB/c mice were infected intranasally with 2 × 105 PFU of MHV-68, MHV-72 and MHV-4556. The lungs were harvested at 10, 14, 21, 35, and 42 dpi and reactivation of virus was induced by the ex vivo explantation of the lungs tissue in the presence of TSA (200 ng/mL) for 10 days. (A) Titer of the infectious virus released from lungs during the explantation at 3, 7, and 10 dpe. The viral titer in the explantation medium was determined by plaque assay on Vero cells. (B) Viral genome copies of MHV-68 vs. MHV-72 vs. MHV-4556 in the lungs tissue at day 10 after the TSA-induced ex vivo reactivation. The number of viral genome copies was determined by qPCR. Data are presented as the mean of PFU/tissue (log10) (A) or genome copy number (log10) (B) ± SEM and represent averages of two independent experiments. Five mice per group were assayed at each time interval. *P ≤ 0.05, **P ≤ 0.01 ***P ≤ 0.001 expresses the statistical significance of differences between reactivation (R) versus latency (L) of individual strains of MHV-68, MHV-72, and MHV-4556; and the statistical significance of differences between reactivated MHV-72 or MHV-4556 versus MHV-68. dpi – day post infection; L – latency (genome copies number detected before induced ex vivo reactivation); R – reactivation (genome copies number detected after induced ex vivo reactivation).
MHV-72 and MHV-4556 have a reduced capacity for ex vivo reactivation in the lungs
At the end of acute infection (10 dpi), MHV-72 showed a high ability to be released from the lungs treated with TSA ex vivo, as evidenced by high titers of de novo assembled infectious virus detected at 3, 7 and 10 dpe (Figure 3A), although copies of the MHV-72 genome were similar to those of MHV-68 (Figure 3B). However, the reactivation ability of MHV-72 continuously decreased with the progression of latency. At 35 and 42 dpi, copies of the MHV-72 genome detected in lungs tissue after TSA-induced reactivation were similar to that detected during viral latency (Figure 3B) and the titers of de novo assembled MHV-72 virus reached the lowest levels–only up to 100 PFU/tissue (Figure 3A). These results show that MHV-72 has a reactivation defect from latency in the lungs.
At the end of acute infection (10 dpi), the ability of MHV-4556 to reactivate from lungs was similar to MHV-68, which was confirmed by the titer of de novo assembled virus detected in medium at 3, 7, and 10 dpe (Figure 3A) and also by the number of MHV-4556 genome copies in the lungs after TSA-induced reactivation (Figure 3B). However, we observed that the ability of MHV-4556 to reactivate from the lungs, similar to that of MHV-72, continuously decreased with the progression of latency, reaching the lowest levels in the number of MHV-4556 genome copies, as well as the titer of reactivated virus at 42 dpi (Figures 3A, B).
Taken together, we found that in addition to its deficiency in latency establishment, MHV-72 has a significantly reduced ability to reactivate from the lungs ex vivo in both early and deep latency. Similarly, MHV-4556 has reduced capacity for ex vivo reactivation from lungs, although its ability to establish latency in lungs is comparable with MHV-68.
Reactivation from spleen
The ability of MHV-72 and MHV-4556 to ex vivo reactivate from spleen in comparison to MHV-68 is demonstrated in Figure 4. MHV-68 showed high ability to reactivate from spleen after the stimulation with TSA. The titer of de novo assembled MHV-68 released into the explantation medium was similar at all time-intervals post infection (10–42 dpi), reaching approximately 3.7 log10 and 5.3 log10 PFU/tissue at 7 and 10 dpe, respectively (Figure 4A). Concurrently, the level of MHV-68 genome copies was about two-times higher in comparison to the level of latent virus detected in spleen immediately after the mice sacrifice (Figure 4B). The ability of MHV-68 to reactivate was continuously increasing with the progression of latency, reaching the highest number of genome copies at 35 dpi, when the deep latency was established.
FIGURE 4
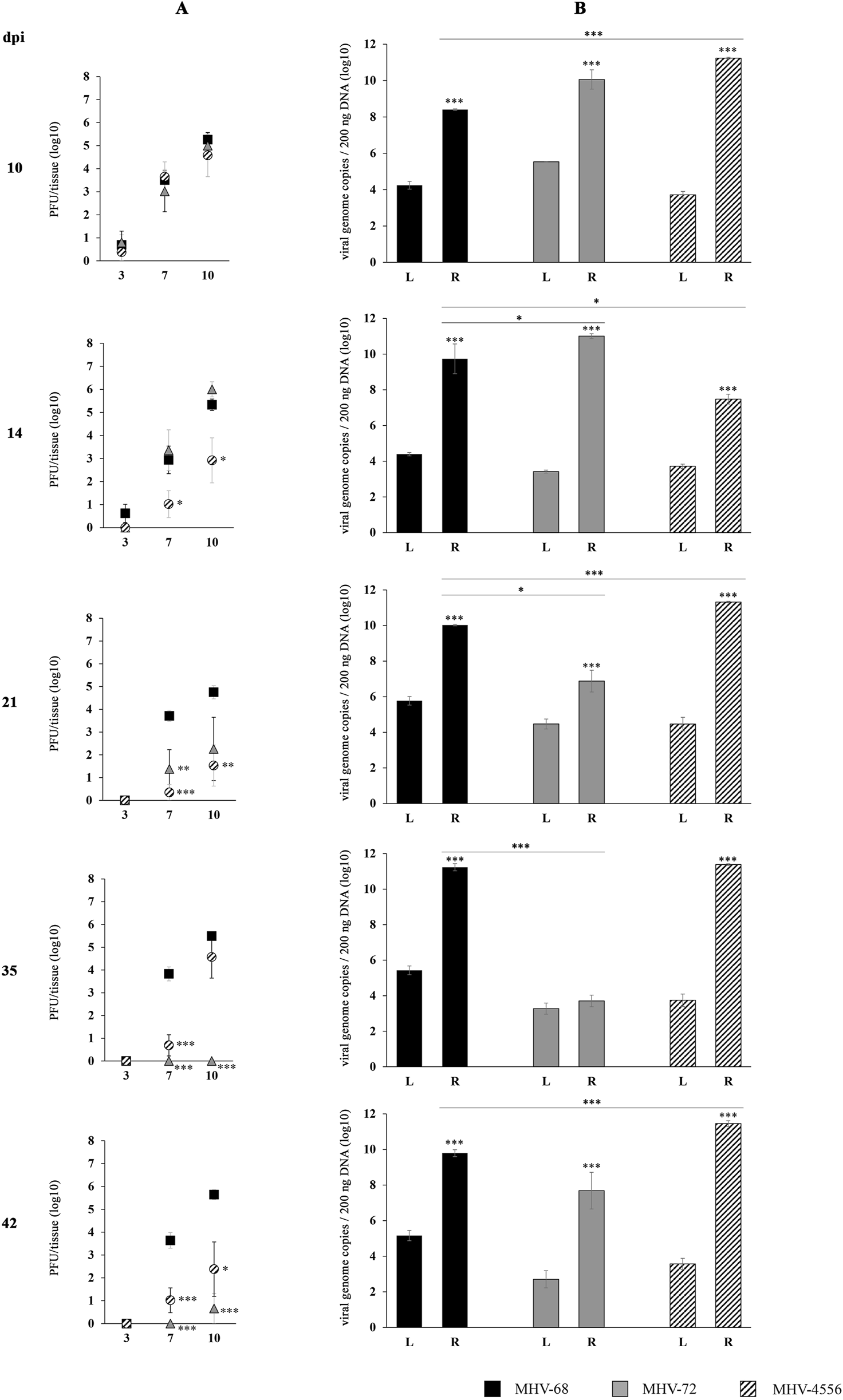
TSA-induced ex vivo reactivation of MHV-72 and MHV-4556 in the spleen. BALB/c mice were infected intranasally with 2 × 105 PFU of MHV-68, MHV-72 and MHV-4556. The spleen was harvested at 10, 14, 21, 35, and 42 dpi and reactivation of virus was induced by the ex vivo explantation of the spleen tissue in the presence of TSA (200 ng/mL) for 10 days. (A) Titer of the infectious virus released from spleen during the explantation at 3, 7, and 10 dpe. The viral titer in the explantation medium was determined by plaque assay on Vero cells. (B) Viral genome copies of MHV-68 vs. MHV-72 vs. MHV-4556 in the spleen tissue at day 10 after the TSA-induced ex vivo reactivation. The number of viral genome copies was determined by qPCR. Data are presented as the mean of PFU/tissue (log10) (A) or genome copy number (log10) (B) ± SEM and represent averages of two independent experiments. Five mice per group were assayed at each time interval. *P ≤ 0.05, **P ≤ 0.01 ***P ≤ 0.001 expresses the statistical significance of differences between reactivation (R) versus latency (L) of individual strains of MHV-68, MHV-72, and MHV-4556; and the statistical significance of differences between reactivated MHV-72 or MHV-4556 versus MHV-68. dpi–day post infection; L–latency (genome copies number detected before induced ex vivo reactivation); R–reactivation (genome copies number detected after induced ex vivo reactivation).
MHV-72 possesses a deficit in the ex vivo reactivation from spleen. MHV-4556 is able to reactivate from latency in spleen, but has a defect in the release from tissue
The titer of de novo assembled MHV-72 detected in medium of explanted spleen at 10 and 14 dpi was comparable to that of MHV-68 (Figure 4A). As we presented in Figure 2, the copy number of the MHV-72 genome was significantly higher than that of the MHV-68 genome prior to induction of reactivation in the spleen at 10 dpi. Similarly, after ex vivo reactivation, MHV-72 genome copy number increased more than MHV-68 copy number at 10 dpi, although not significantly (Figure 4B). This increase in MHV-72 genome copy number in spleen tissue after TSA-induced reactivation was maintained at 14 dpi. However, from 21 to 42 dpi, the genome copy number as well as the reactivated virus titers of MHV-72 decreased significantly compared with MHV-68. The lowest number of MHV-72 genome copies was detected at 35 dpi, app. 3.7 log10 genome copies of MHV-72 compared to 11.23 log10 genome copies of MHV-68 (P ≤ 0.001) (Figure 4B). Similarly, the level of the reactivated MHV-72 released into the explantation medium was almost not-detectable at 35 and 42 dpi (Figure 4A). Therefore, we can conclude that MHV-72 is deficient not only in establishment of latency in the spleen but also in reactivation from it.
The course of MHV-4556 reactivation from the spleen was different in comparison to MHV-72 as well as MHV-68 (Figure 4). Although MHV-4556 genome copy number before induction of reactivation was equal (10 dpi) or lower (14–42 dpi) than MHV-68 copy number, after induction of reactivation with TSA, MHV-4556 copy number in spleen tissue was at all studied days after infection, except at 14 dpi, higher than MHV-68 (Figure 4B). The highest number of MHV-4556 genome copies was detected at 42 dpi (11.4 log10 MHV-4556 genome copies vs. 9.7 log10 MHV-68 genome copies). In contrast, the titer of de novo assembled MHV-4556, which we detected in the medium at 7 and 10 dpe, decreased with the progression of latency and reached lower levels in comparison to that of MHV-68 (Figure 4A).
Therefore, we can conclude that MHV-4556 is able to reactivate from latency in spleen, however, has a defect in the release from latently infected spleen as a de novo assembled virus.
Reactivation from thymus
The ability of MHV-72 and MHV-4556 to reactivate ex vivo from thymus in comparison to MHV-68 is demonstrated in Figure 5. MHV-68 showed high ability to reactivate from thymus after the stimulation with TSA. The titer of de novo assembled MHV-68 released into the explantation medium at 7 and 10 dpe was increasing up to 21 dpi and then slightly decreased (Figure 5A). The MHV-68 genome copy number in the thymus after TSA-stimulated reactivation was significantly higher than in the latent tissue before reactivation, already reaching high values of 9.2 log10 genome copies at 10 dpi, and this number slowly increased to 10.4 log10 genome copies at 21 dpi. However, with the progression of latency, the number of genome copies after reactivation was slightly decreasing up to 7.9 log10 at 42 dpi (Figure 5B).
FIGURE 5
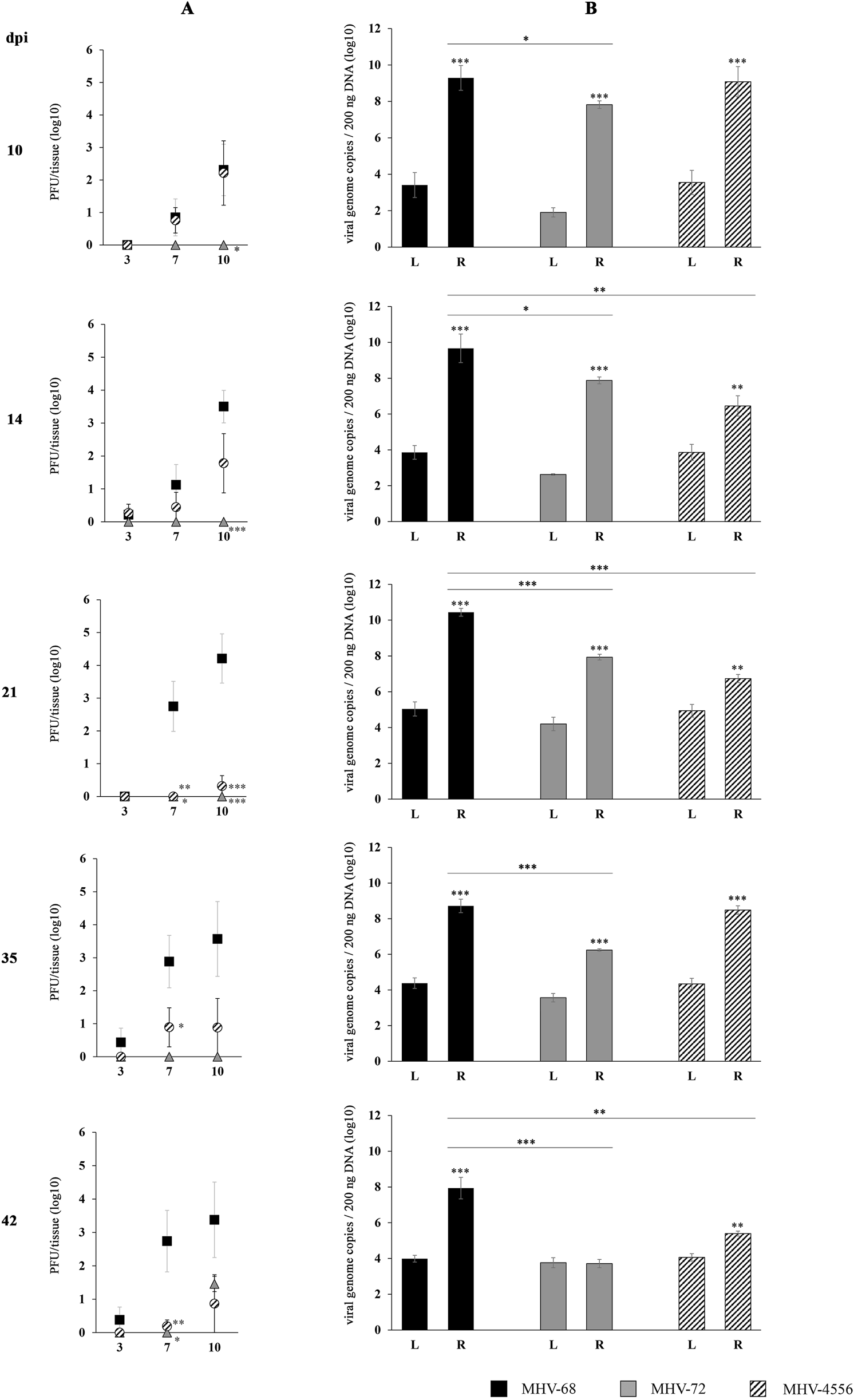
TSA-induced ex vivo reactivation of MHV-72 and MHV-4556 in thymus. BALB/c mice were infected intranasally with 2 × 105 PFU of MHV-68, MHV-72 and MHV-4556. Thymus was harvested at 10, 14, 21, 35, and 42 dpi and reactivation of virus was induced by the ex vivo explantation of the thymus tissue in the presence of TSA (200 ng/mL) for 10 days. (A) Titer of the infectious virus released from thymus during the explantation at 3, 7, and 10 dpe. The viral titer in the explantation medium was determined by plaque assay on Vero cells. (B) Viral genome copies of MHV-68 vs. MHV-72 vs. MHV-4556 in thymus tissue at day 10 after the TSA-induced ex vivo reactivation. The number of viral genome copies was determined by qPCR. Data are presented as the mean of PFU/tissue (log10) (A) or genome copy number (log10) (B) ± SEM and represent averages of two independent experiments. Five mice per group were assayed at each time interval. *P ≤ 0.05, **P ≤ 0.01 ***P ≤ 0.001 expresses the statistical significance of differences between reactivation (R) versus latency (L) of individual strains of MHV-68, MHV-72, and MHV-4556; and the statistical significance of differences between reactivated MHV-72 or MHV-4556 versus MHV-68. dpi–day post infection; L–latency (genome copies number detected before induced ex vivo reactivation); R–reactivation (genome copies number detected after induced ex vivo reactivation).
MHV-72 has significantly reduced capacity for ex vivo reactivation from thymus. MHV-4556 has a reduced capacity for ex vivo reactivation from thymus only during the deep latency
The capacity of MHV-72 for reactivation from thymus was significantly lower in comparison to MHV-68 (Figures 5A, B). Although, we detected slightly increased titer of MHV-72 at 42 dpi (1.46 log10 PFU/tissue at 10 dpe), at 10–35 dpi we observed the inability of MHV-72 to release from thymus when titer of de novo assembled MHV-72 was almost non-detectable (Figure 5A). Similarly, although TSA treatment induced an increase in MHV-72 genome copy number compared to pre-reactivation levels at 10–35 dpi, the copy number of reactivated MHV-72 was, however, significantly lower compared to MHV-68 at all examined dpi. And finally, the lowest level of MHV-72 genome copies we detected at 42 dpi (3.7 log10 MHV-72 genome copies vs. 7.9 log10 MHV-68 genome copies, P ≤ 0.001) (Figure 5B).
We also found a reduced ability of MHV-4556 to reactivate from the thymus (Figures 5A, B). At 10 dpi and 35 dpi, the number of MHV-4556 genome copies after TSA-induced reactivation was similar to MHV-68. However, at 14, 21, and 42 dpi, the number of MHV-4556 genome copies was decreased by about 3.0–3.5 log10 (P ≤ 0.001 and P ≤ 0.01). Reduced ability of MHV-4556 to reactivate from thymus during the deep latency was also confirmed by low levels of titer of infectious virus detected at 14–42 dpi (Figure 5A) (decreased by about 2.5–4 log10 (P ≤ 0.001 and P ≤ 0.01) PFU/tissue in comparison to MHV-68).
These results suggest that both strains, MHV-72 and MHV-4556, have a significantly reduced ability to reactivate ex vivo from the thymus compared to MHV-68.
Discussion
It is known that the primary infection of human gammaherpesviruses is often asymptomatic or accompanied by benign cellular proliferation in the initial site of entry but finally results in lifelong latent infection. However, the latency of human gammaherpesviruses can lead to development of lymphoproliferative diseases and malignancies (Chinna et al., 2023). Despite the fact that most of the EBV and KSHV strains do not differ from each other in the course of acute infection, several mutations were found mostly in genes responsible for maintaining latency and reactivation. It was found that differences identified in a small subset of the EBV-encoded latent genes, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C and LMP-1 vary markedly according to not only geographical location but also within individual Burkitt’s lymphoma tumors (Palser et al., 2015; Xie et al., 2024). Similarly, geographical variation in KSHV genes may relate to differences in virulence or transmission (Olp et al., 2015; Sallah et al., 2020).
Therefore, we aimed to analyze the strain-related variations in the latency and reactivation of MuHV-4 (alias MHV-68), which is commonly used as a model to study of human gammaherpesviruses (Fujiwara and Nakamura, 2020). Here, we quantified the number of viral genome copies of three strains/isolates or genotype variants of MuHV-4, MHV-68 (as a prototype), MHV-72 (clone h3.7) and MHV-4556 (clone i2.8) in lungs, spleen and thymus during the course of latency establishment (at 10, 14, 21, 35, and 42 dpi) as well as ex vivo reactivation induced by TSA treatment of tissues during the explantation procedure (Figure 6). This approach represents the important contribution to the molecular characterization of pathogenic differences among the strains/isolates or genotype variants of MuHV-4.
FIGURE 6
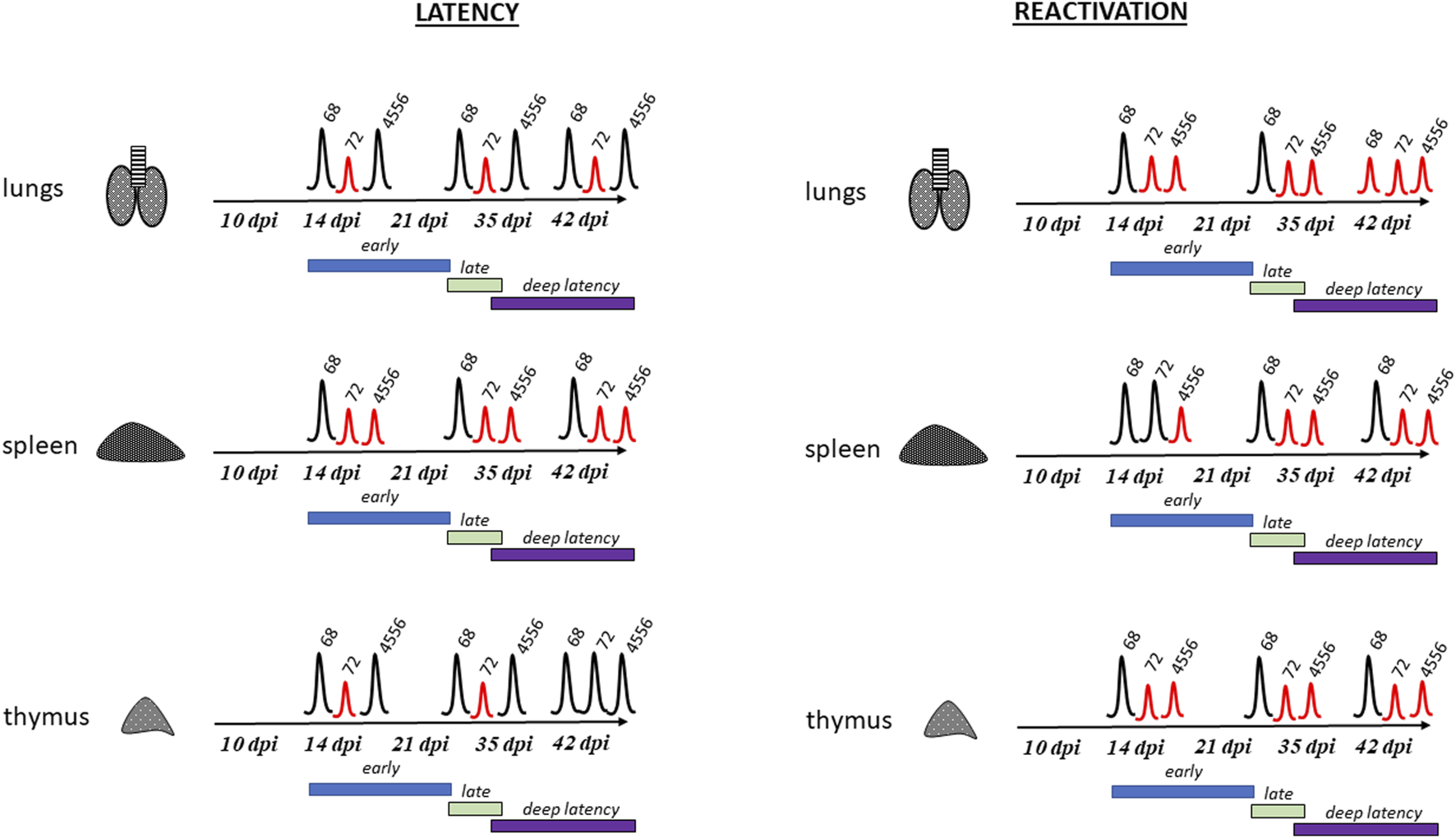
Major differences between MHV-68, MHV-72 and MHV-4556 latency and reactivation. Left panel shows latency establishment of strain MHV-68, MHV-72, and MHV-4556 in lungs, spleen and thymus of infected mice. Right panel shows ability of strain MHV-68, MHV-72, and MHV-4556 to ex vivo reactivate from lungs, spleen and thymus. Time course of early, late and deep latency is shown in scheme. Peaks show ability of virus to establish latency and reactivate, red peaks show significant differences of strains MHV-72 or MHV-4556 from WT MHV-68.
Similarly to human gammaherpesviruses, MuHV-4 latency occurs mainly in B cells of germinal centers and marginal zone B cells of spleen. However, the latency of MHV-68 can be established also in T cells, natural killer cells, surface immunoglobulin M-negative pre-B cells, bone marrow pro-pre-B cells, immature B cells, peritoneal macrophages, epithelial cells of thymus, pulmonary epithelial cells, lymph nodes as well as salivary glands (Coleman et al., 2010; Barton et al., 2011; Riggs et al., 2021; Wang et al., 2021). Additional sites, such as urogenital tract, tail and peritoneal cavity were found to be positive for MHV-68 at later times post infection (Hwang et al., 2008). The latent infection can be established very early during the acute response, the B cells in lungs harbor latent virus as early as 3 days after infection. Latency in spleen can be detected as soon as 10–18 dpi, however, the general rule is that the early latency is not recognized earlier than 15 dpi and late latency is established at 42 dpi and later (Johnson and Tarakanova, 2020; Zelazowska et al., 2020; Wang et al., 2021). The persistent infection of MHV-68 in lungs can be detected even at 90 dpi and the lymphoid infiltrate can persists in lungs up to 170 dpi (Sunil-Chandra et al., 1992; Stevenson and Doherty, 1998). Therefore, it is not surprising that the virus persisting in the lungs could seed the spleen, and conversely, virus resident in the spleen could seed the lungs during the long-term persistent infection (Stewart et al., 1998).
In order to analyze strain-dependent differences in latency, we compared the time course and level of latency establishment of MHV-72 (clone h3.7) and MHV-4556 (clone i2.8) with prototype strain MHV-68. In the lungs, spleen and thymus of mice intranasally infected with MHV-68, MHV-72 or MHV-4556, we assessed the amount of infectious virus by ICA and the number of viral genome copies by qPCR at five intervals - 10, 14, 21, 35, and 42 dpi (Figure 1). We found no significant differences in the amount of infectious virus present in the lung, spleen, and thymus among the viral strains. Only low level of infectious virus was detected, reaching a maximum of 100 PFU in the lungs at 10 dpi. Later, from 14 to 42 dpi, infectious virus was almost undetectable in all organs tested (max. 20 PFU/tissue). Therefore, we hypothesize that the copies of the viral genome that we detected in the organs examined are likely to be copies of the genome of a non-infectious, latent virus. And in this parameter, the individual viral strains were already significantly different from each other. We detected a reduced ability of MHV-72 and MHV-4556 to establish latency, which could be observed mostly in spleen. More specifically, with the exception of 10 dpi (the end of acute infection), the number of MHV-72 genome copies in spleen, lungs, and thymus was lower in comparison to MHV-68 during the early (from 14 dpi) as well as deep (from 35 dpi) latency (Figure 2). In comparison to this, the latency establishment of MHV-4556 in lungs and thymus was similar to MHV-68 at all-time points. However, in spleen, we detected significantly decreased ability of MHV-4556 to establish latency from 14 dpi up to 42 dpi (Figure 2). Similar observation was made also by Macrae et al. 2001, when MHV-76, another strain of MuHV-4, was able to establish latency in spleen during the first 30 days of infection, albeit at a lower level than MHV-68. MHV-76 genome is essentially identical to that of MHV-68, except for a 9,538-bp deletion at the left end of the unique region, involving genes M1, M2, M3, M4, and all of the vtRNA genes. Currently, the whole genome sequencing revealed deletion in the left end of MHV-72 (clone h3.7) and also MHV-4556 (clone i2.8) genomes, causing the absence of four vtRNAs, M1, and M2 genes at least. Earlier, the different amino-acid sequences of proteins encoded by MHV-72 (clone h3.7) were found in structural proteins ORF20, ORF26, ORF48, M7 and ORF52 and non-structural proteins M3, ORF4 and MK3 (Belvončíková et al., 2008; Halásová et al., 2011). Moreover, the different amino-acid sequences of proteins encoded by MHV-4556 (clone i2.8) were found in structural proteins ORF8, ORF11, ORF26, ORF48 and ORF52 and non-structural proteins M4 (Kúdelová et al., 2012) and MK3 (Valovičová et al., 2006). Similar to MHV-76 (Macrae et al., 2001), we hypothesize that the reduced ability of MHV-72 and MHV-4556 to establish latency in spleen could be caused mainly by deletion of genes at the left-end of the genome and subsequent inability to properly interact with host system (see below).
Using the explantation procedure (Rajcani et al., 1985) in combination with treatment by a histone deacetylase inhibitor trichostatin A (Yang et al., 2009) we found that all three strains were able to reactivate from latency into relatively high levels from spleen at all tested intervals. However, the reactivation rate of MHV-72 and MHV-4556, as measured by the number of the viral genome copies in the tissue after the reactivation procedure (explantation with TSA treatment), was lower compared with MHV-68. More specifically, in addition to the MHV-72 deficiency in establishment of latency in lungs, we identified its significantly reduced capacity for ex vivo reactivation from lungs at early latency (14–21 dpi) and incapability to reactivate from deep latency (35–42 dpi) (Figure 3). Similarly, MHV-72 has a deficit in the ex vivo reactivation from spleen after 21 dpi (Figure 4), and also from thymus, which was detected mostly at 35 and 42 dpi (Figure 5). Moreover, we observed the inability of MHV-72 to release from explanted tissue of thymus, when the titer of de novo assembled MHV-72 was almost non-detectable at 10–35 dpi. Taken together, MHV-72 exhibited the significant deficit not only in the establishment of latency in all studied organs but also in ex vivo reactivation stimulated by TSA. In contrast, MHV-4556 was able to respond to the TSA-mediated reactivation stimuli by increasing the DNA copy number but had a defect in the release from tissue during the early and deep latency in spleen (Figure 4). However, MHV-4556 exhibited reduced capacity for ex vivo reactivation from early as well as deep latency in lungs (Figure 3) and also significantly reduced capacity for ex vivo reactivation at deep latency in thymus (Figure 5).
In spite of the very effective reactivation of MHV-68 induced by TSA in all studied organs, the capacity for its reactivation slightly decreased during the deep latency, at 35 and 42 dpi. Similarly to this observation, previously it was found, that MHV-68 reactivates efficiently from latently infected cells isolated early after the mice infection. However, with progression of latency, the efficiency of reactivation decreases (Weck et al., 1999; Tibbetts et al., 2003).
Despite the intensive study, mechanisms leading to gammaherpesvirus reactivation from latency are not very well understood. It is known that Rta (encoded by ORF50) is a mediator of molecular switch and activates the expression of early (lytic) genes, therefore, plays a key role in virus reactivation from latency (Wu et al., 2000; Pavlova et al., 2003). In previous study, we did not find any differences in nucleotide sequences of Rta of MHV-68, MHV-72 and MHV-4556, as well as any differences in the epigenetic regulation of Rta promoter during the lytic replication, latency and also ex vivo and in vivo reactivation from spleen induced by TSA (Lapuníková et al., 2015). It follows that the unique in vivo pathogenic properties of MHV-72 (clone h3.7) and MHV-4556 (clone i2.8), which are identified in this study, are not related to the functional activity of Rta and Rta-induced molecular switch.
How can we explain the different ability of MuHV-4 strains to establish latency in vivo and to reactivate ex vivo? As we mentioned above, the one explanation may be related to the differences in primary genome structure of MuHV-4 strains. Mainly the M2 gene seems to be the first candidate for sufficient explanation. It was demonstrated that M2 protein plays a critical role in both the establishment of latency as well as virus reactivation from latently infected B cells, although is completely dispensable for virus replication in permissive cell lines, as well as in the lungs following intranasal inoculation (Owens et al., 2020; Wang et al., 2021). Analyses of virus latency have shown that MHV-68 is present in naive, germinal center and memory B cells of spleen during the early latency. However, latency in naive and germinal center B cells wanes over time and is predominantly found in memory B cells at late times post infection, which are then rapidly differentiated into plasma cells (Terrell and Speck, 2017; Wang et al., 2021). In connection with that (Liang et al. 2009), showed that in mice infected with wild type MHV-68, virus-infected plasma cells (cca 8% from all of the virus-infected splenocytes at the peak of viral latency) are the source of majority of reactivated virus observed upon explantation of splenocytes. In mice infected with a M2-null MHV-68, virus-infected plasma cells are completely missing at the peak of latency. In addition, infection with M2-null virus leads to a more rapid clearance of replication in the spleen following high dose intraperitoneal inoculation and this mutant virus is not able to reactivate. Furthermore (Liang et al. 2009), clearly demonstrated that M2 protein is responsible for terminal differentiation of B cells to plasma cells in vivo.
In addition to the important role of M2 protein in differentiation of B cells, it was shown, that M2 protein is expressed in different cell types during the latent infection of MHV-68 and antagonizes IFN-α/β- and IFN-γ-mediated transcriptional activation. In lymphocytes, M2 protein is present in the cytoplasm and plasma membrane, and only in low amount in nucleus, whereas in fibroblast, epithelial, and endothelial cells, it is present primarily in the nucleus, and in a low amount in the cytoplasm and plasma membrane (Liang et al., 2004). Thus, M2 represents a new class of herpesvirus gene products (reactivation conditioners) that do not directly participate in virus replication, but rather facilitate virus reactivation by manipulating the cellular milieu to provide a reactivation competent environment. Results presented in this work demonstrate the reduced ability of two strains of MuHV-4, MHV-72 and MHV-4556, not only to establish latency in vivo, but also to reactivate ex vivo. Given that both MHV-72 (clone h3.7) and MHV-4556 (clone i2.8) do not contain the M2 gene in their genomes, it is highly probable that this is the main cause of their reduced ability to establish latency in comparison to prototype MHV-68. However, to confirm the foregoing, it would be appropriate to evaluate the MHV-72 and MHV-4556 latency establishment in the individual subpopulation of B cells as well as to identify the differentiation status of B cells harboring the MuHV-4 genome.
Nevertheless, only the absence of M2 gene in genomes of both MHV-72 and MHV-4556 do not explain their decreased ability to reactivate from latency and, unfortunately, currently we are not able to sufficiently resolve the differences among these two strains. Therefore, the future goal might be to confirm our findings on the strain-dependent reactivation ability in vivo, mainly in the context of M2 deficiency in MHV-72 and MHV-4556 genomes.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by The State Veterinary and Food Administration of the Slovak Republic under permit number Ro. 3497/14-221. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
KL, PBe, IS, VV, PBa, and MBe conducted the experiments. TS and MBö conducted bioinformatical analysis. KL and IR wrote the manuscript, and MK contributed with valuable comments and advice. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the projects VEGA #2/0185/12 and VEGA #2/0063/21 funded by the Scientific Grant Agency of Slovak Ministry of Education and Slovak Academy of Sciences, by the project 101160008 — FORGENOM II — HORIZON-WIDERA-2023-ACCESS-02 funded by the Horizon Europe programme, and by the Slovak Research and Development Agency grant APVV-20-0472 (Sepmin).
Conflict of interest
Authors MBö and TS were employed by the company Geneton s.r.o.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Abbreviations
dpe, day post explantation; dpi, day post infection; EBV, Epstein-Barr virus; ICA, infectious centre assay; KSHV, Kaposi’s sarcoma-associated herpesvirus; MHV-68, Murine gammaherpesvirus 68; MuHV-4, Murid gammaherpesvirus 4; MOI, multiplicity of infection; SCS, single cell suspension; TSA, trichostatin A.
References
1
Adler H. Messerle M. Wagner M. Koszinowski U. H. (2000). Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol.74, 6964–6974. 10.1128/JVI.74.15.6964-6974.2000
2
Barton E. Mandal P. Speck S. H. (2011). Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu. Rev. Immunol.29, 351–397. 10.1146/annurev-immunol-072710-081639
3
Belvončíková P. Kráľová A. Kúdelová M. Hajnická V. Režuchová I. Vančová I. (2008). Chemokine-binding activities of M3 protein encoded by murine gammaherpesvirus 72. Acta Virol.52, 91–97.
4
Blaskovic D. Mistrikova J. (1985). Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol.29, 312–317.
5
Blaskovic D. Stancekova M. Svobodova J. Mistrikova J. (1980). Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol.24, 468.
6
Buschle A. Hammerschmidt W. (2020). Epigenetic lifestyle of Epstein-Barr virus. Semin. Immunopathol.42, 131–142. 10.1007/s00281-020-00792-2
7
Chinna P. Bratl K. Lambarey H. Blumenthal M. Schäfer G. (2023). The impact of Co-infections for human gammaherpesvirus infection and associated pathologies. IJMS24, 13066. 10.3390/ijms241713066
8
Čipková-Jarčušková J. Chalupková A. Hrabovská Z. Wágnerová M. Mistríková J. (2013). Biological and pathogenetic characterization of different isolates of murine gammaherpesvirus 68 (MHV-68) in the context of study of human oncogenic gammaherpesviruses. Acta Virol.57, 105–112. 10.4149/av_2013_02_105
9
Coleman C. B. Nealy M. S. Tibbetts S. A. (2010). Immature and transitional B cells are latency reservoirs for a gammaherpesvirus. J. Virol.84, 13045–13052. 10.1128/jvi.01455-10
10
de Martel C. Georges D. Bray F. Ferlay J. Clifford G. M. (2020). Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health8 (2), e180–e190. 10.1016/S2214-109X(19)30488-7
11
Frederico B. Milho R. May J. S. Gillet L. Stevenson P. G. (2012). Myeloid infection links epithelial and B cell tropisms of murid herpesvirus-4. PLOS Pathog.8, e1002935. 10.1371/journal.ppat.1002935
12
Fujiwara S. Nakamura H. (2020). Animal models for gammaherpesvirus infections: recent development in the analysis of virus-induced pathogenesis. Pathogens9, 116. 10.3390/pathogens9020116
13
Guo H. Wang L. Peng L. Zhou Z. H. Deng H. (2009). Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J. Virol.83, 10582–10595. 10.1128/JVI.00497-09
14
Halásová Z. Valovičová M. Mačáková K. Pančík P. Belvončíková P. Režuchová I. et al (2011). Partial genome sequence of murine gammaherpesvirus 72 and its analysis. av55, 317–325. 10.4149/av_2011_04_317
15
Hwang S. Wu T.-T. Tong L. M. Kim K. S. Martinez-Guzman D. Colantonio A. D. et al (2008). Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. J. Virol.82, 12498–12509. 10.1128/JVI.01152-08
16
Jary A. Veyri M. Gothland A. Leducq V. Calvez V. Marcelin A.-G. (2021). Kaposi’s sarcoma-associated herpesvirus, the etiological agent of all epidemiological forms of kaposi’s sarcoma. Cancers13, 6208. 10.3390/cancers13246208
17
Johnson K. E. Tarakanova V. L. (2020). Gammaherpesviruses and B Cells: a relationship that lasts a lifetime. Viral Immunol.33, 316–326. 10.1089/vim.2019.0126
18
Kozuch O. Reichel M. Lesso J. Remenova A. Labuda M. Lysy J. et al (1993). Further isolation of murine herpesviruses from small mammals in southwestern Slovakia. Acta Virol.37, 101–105.
19
Kúdelová M. Halásová Z. Belvončíková P. Pančík P. Režuchová I. Valovičová M. (2012). Partial genome analysis of murine gammaherpesvirus 4556. av56, 177–186. 10.4149/av_2012_03_177
20
Lapuníková B. Lopušná K. Benkóczka T. Golais F. Kúdelová M. Režuchová I. (2015). Epigenetic modification of Rta (ORF50) promoter is not responsible for distinct reactivation patterns of murine gammaherpesviruses. av59, 405–412. 10.4149/av_2015_04_405
21
Liang X. Collins C. M. Mendel J. B. Iwakoshi N. N. Speck S. H. (2009). Gammaherpesvirus-driven plasma cell differentiation regulates virus reactivation from latently infected B lymphocytes. PLoS Pathog.5, e1000677. 10.1371/journal.ppat.1000677
22
Liang X. Shin Y. C. Means R. E. Jung J. U. (2004). Inhibition of interferon-mediated antiviral activity by murine gammaherpesvirus 68 latency-associated M2 protein. J. Virol.78, 12416–12427. 10.1128/JVI.78.22.12416-12427.2004
23
Macrae A. I. Dutia B. M. Milligan S. Brownstein D. G. Allen D. J. Mistrikova J. et al (2001). Analysis of a novel strain of murine gammaherpesvirus reveals a genomic locus important for acute pathogenesis. J. Virol.75, 5315–5327. 10.1128/JVI.75.11.5315-5327.2001
24
Mistríková J. Briestenská K. (2020). Murid herpesvirus 4 (MuHV-4, prototype strain MHV-68) as an important model in global research of human oncogenic gammaherpesviruses. Acta Virol.64, 167–176. 10.4149/av_2020_206
25
Nehme Z. Pasquereau S. Herbein G. (2020). “Targeting histone epigenetics to control viral infections,” in Histone modifications in therapy (Elsevier), 255–292. 10.1016/B978-0-12-816422-8.00011-8
26
Olp L. N. Jeanniard A. Marimo C. West J. T. Wood C. (2015). Whole-genome sequencing of kaposi’s sarcoma-associated herpesvirus from Zambian kaposi’s sarcoma biopsy specimens reveals unique viral diversity. J. Virology89, 12299–12308. 10.1128/jvi.01712-15
27
Owens S. M. Oldenburg D. G. White D. W. Forrest J. C. (2020). Deletion of murine gammaherpesvirus gene M2 in activation-induced cytidine deaminase-expressing B cells impairs host colonization and viral reactivation. J. Virol.95, 019333–20. 10.1128/JVI.01933-20
28
Palser A. L. Grayson N. E. White R. E. Corton C. Correia S. Ba Abdullah M. M. et al (2015). Genome diversity of epstein-barr virus from multiple tumor types and normal infection. J. Virol.89, 5222–5237. 10.1128/JVI.03614-14
29
Pavlova I. V. Virgin H. W. Speck S. H. (2003). Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J. Virol.77, 5731–5739. 10.1128/JVI.77.10.5731-5739.2003
30
Rajcani J. Blaskovic D. Svobodova J. Ciampor F. Huckova D. Stanekova D. (1985). Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol.29, 51–60.
31
Raslova H. Mistrikova J. Kudelova M. Mishal Z. Sarasin A. Blangy D. et al (2000). Immunophenotypic study of atypical lymphocytes generated in peripheral blood and spleen of nude mice after MHV-72 infection. Viral Immunol.13, 313–327. 10.1089/08828240050144644
32
Riggs J. B. Medina E. M. Perrenoud L. J. Bonilla D. L. Clambey E. T. Van Dyk L. F. et al (2021). Optimized detection of acute MHV68 infection with a reporter system identifies large peritoneal macrophages as a dominant target of primary infection. Front. Microbiol.12, 656979. 10.3389/fmicb.2021.656979
33
Sallah N. Miley W. Labo N. Carstensen T. Fatumo S. Gurdasani D. et al (2020). Distinct genetic architectures and environmental factors associate with host response to the γ2-herpesvirus infections. Nat. Commun.11, 3849. 10.1038/s41467-020-17696-2
34
Shareena G. Kumar D. (2023). Epigenetics of Epstein barr virus — a review. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis.1869, 166838. 10.1016/j.bbadis.2023.166838
35
Stevenson P. G. Doherty P. C. (1998). Kinetic analysis of the specific host response to a murine gammaherpesvirus. J. Virol.72, 943–949. 10.1128/JVI.72.2.943-949.1998
36
Stewart J. P. Usherwood E. J. Ross A. Dyson H. Nash T. (1998). Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med.187, 1941–1951. 10.1084/jem.187.12.1941
37
Su Z. Y. Siak P. Y. Leong C.-O. Cheah S.-C. (2023). The role of Epstein–Barr virus in nasopharyngeal carcinoma. Front. Microbiol.14, 1116143. 10.3389/fmicb.2023.1116143
38
Sunil-Chandra N. P. Efstathiou S. Nash A. A. (1992). Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. General Virology73, 3275–3279. 10.1099/0022-1317-73-12-3275
39
Terrell S. Speck S. H. (2017). Murine gammaherpesvirus M2 antigen modulates splenic B cell activation and terminal differentiation in vivo. PLoS Pathog.13, e1006543. 10.1371/journal.ppat.1006543
40
Tibbetts S. A. Loh J. Van Berkel V. McClellan J. S. Jacoby M. A. Kapadia S. B. et al (2003). Establishment and maintenance of gammaherpesvirus latency are independent of infective dose and route of infection. J. Virol.77, 7696–7701. 10.1128/JVI.77.13.7696-7701.2003
41
Tsuji N. Kobayashi M. Nagashima K. Wakisaka Y. Koizumi K. (1976). A new antifungal antibiotic, trichostatin. J. Antibiot.29, 1–6. 10.7164/antibiotics.29.1
42
Valovičová M. Režuchová I. Mačáková K. Petrová P. Matis J. Kúdelová M. (2006). Murine gammaherpesvirus (MHV) MK3 gene sequence diversity among 72, 4556, and 68 strains. Virus Genes33, 51–58. 10.1007/s11262-005-0038-4
43
Wang Y. Tibbetts S. A. Krug L. T. (2021). Conquering the host: determinants of pathogenesis learned from murine gammaherpesvirus 68. Annu. Rev. Virol.8, 349–371. 10.1146/annurev-virology-011921-082615
44
Weck K. E. Barkon M. L. Yoo L. I. Speck S. H. Virgin Hw I. V. (1996). Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol.70, 6775–6780. 10.1128/jvi.70.10.6775-6780.1996
45
Weck K. E. Kim S. S. Virgin H. W. Speck S. H. (1999). B cells regulate murine gammaherpesvirus 68 latency. J. Virol.73, 4651–4661. 10.1128/JVI.73.6.4651-4661.1999
46
Wu T.-T. Usherwood E. J. Stewart J. P. Nash A. A. Sun R. (2000). Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol.74, 3659–3667. 10.1128/JVI.74.8.3659-3667.2000
47
Xie R. Cao B. Wu Z. Ouyang Y. Chen H. Zhai W. et al (2024). dbEBV: a database of Epstein-Barr virus variants and their correlations with human health. Comput. Struct. Biotechnol. J.23, 2076–2082. 10.1016/j.csbj.2024.04.043
48
Yang Z. Tang H. Huang H. Deng H. (2009). RTA promoter demethylation and histone acetylation regulation of murine gammaherpesvirus 68 reactivation. PLoS ONE4, e4556. 10.1371/journal.pone.0004556
49
Zelazowska M. A. McBride K. Krug L. T. (2020). Dangerous liaisons: gammaherpesvirus subversion of the immunoglobulin repertoire. Viruses12, 788. 10.3390/v12080788
Summary
Keywords
murine gammaherpesvirus, MuHV-4, MHV-68, MHV-72, MHV-4556
Citation
Lopusna K, Belvoncikova P, Stibraniova I, Bartikova P, Valentinova V, Benko M, Kudelova M, Böhmer M, Szemes T and Rezuchova I (2025) Murid gammaherpesvirus 4 establishes latency and reactivates in a strain-dependent manner. Acta Virol. 69:13909. doi: 10.3389/av.2025.13909
Received
09 October 2024
Accepted
05 March 2025
Published
25 March 2025
Volume
69 - 2025
Edited by
Ivan Kosik, National Institute of Allergy and Infectious Diseases (NIH), United States
Reviewed by
Jaroslav Holly, National Institute of Allergy and Infectious Diseases (NIH), United States
Updates
Copyright
© 2025 Lopusna, Belvoncikova, Stibraniova, Bartikova, Valentinova, Benko, Kudelova, Böhmer, Szemes and Rezuchova.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katarina Lopusna, katarina.lopusna@savba.sk
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.