Abstract
Alcohol use disorder (AUD) has a complex, multifactorial etiology involving dysregulation across several brain regions and peripheral organs. Acute and chronic alcohol consumption cause epigenetic modifications in these systems, which underlie changes in gene expression and subsequently, the emergence of pathophysiological phenotypes associated with AUD. One such epigenetic mechanism is methylation, which can occur on DNA, histones, and RNA. Methylation relies on one carbon metabolism to generate methyl groups, which can then be transferred to acceptor substrates. While DNA methylation of particular genes generally represses transcription, methylation of histones and RNA can have bidirectional effects on gene expression. This review summarizes one carbon metabolism and the mechanisms behind methylation of DNA, histones, and RNA. We discuss the field’s findings regarding alcohol’s global and gene-specific effects on methylation in the brain and liver and the resulting phenotypes characteristic of AUD.
Introduction
The development of alcohol use disorder (AUD) involves a self-perpetuating cycle of drinking and withdrawal. When an individual consumes alcohol, their reward pathways are stimulated, and they experience reductions in anxiety (1, 2). Chronic alcohol users suffer from cravings and elevated anxiety during withdrawal but can attenuate these symptoms by further drinking (2,3). Many of these behavioral patterns of alcohol consumption and withdrawal, as well as others implicated in the phenotypes of intoxication and addiction, occur as the result of epigenetic changes induced by alcohol (3,4). Particularly, methylation of DNA, histones, and possibly RNA have implications in the underlying pathophysiology of AUD. In this review, we provide an overview of one-carbon metabolism and the process of methylation itself. We discuss and integrate the field’s findings regarding alcohol’s global and gene-specific effects on DNA and histone methylation in both the liver and the brain and how these changes underlie the pathophysiology of AUD (4). Finally, we provide a discussion of RNA methylation, which remains understudied in alcohol addiction. By collectively examining the field’s current knowledge of RNA methylation in conjunction with DNA and histone methylation, we hope this review will provide several potential research avenues on how alcohol consumption affects genomic methylation mechanisms that underlie the neurobiology of AUD.
One-carbon metabolism and regulation of target methylation
Methylation depends on the availability of S-adenosylmethionine (SAM), the universal methyl group donor. SAM is generated by the methionine cycle, a process central to one-carbon metabolism (5). In this cycle, methionine adenosyltransferase (MAT) adenylates methionine using ATP, producing SAM (5–7). SAM contains a methyl group that can be transferred to an acceptor substrate in methylation reactions, leaving S-adenosylhomocysteine (SAH) (5). SAH hydrolase (AHCY) then reversibly converts SAH into adenine and homocysteine (5, 7). However, because the reverse reaction is thermodynamically favored, adenine and homocysteine must be efficiently removed for the reaction kinetics to favor the forward reaction (5–9). The primary process for the removal of these products is the folate cycle, in which dietary folate is used to convert homocysteine into methionine, the precursor of SAM (5–7) (Figure 1A). Betaine can also be used to generate methionine from homocysteine in a process limited to the liver and kidneys (5, 6). Alternatively, the transsulfuration pathway, which is also very active in the liver, removes homocysteine without regenerating methionine (5, 9) (Figure 1B).
FIGURE 1
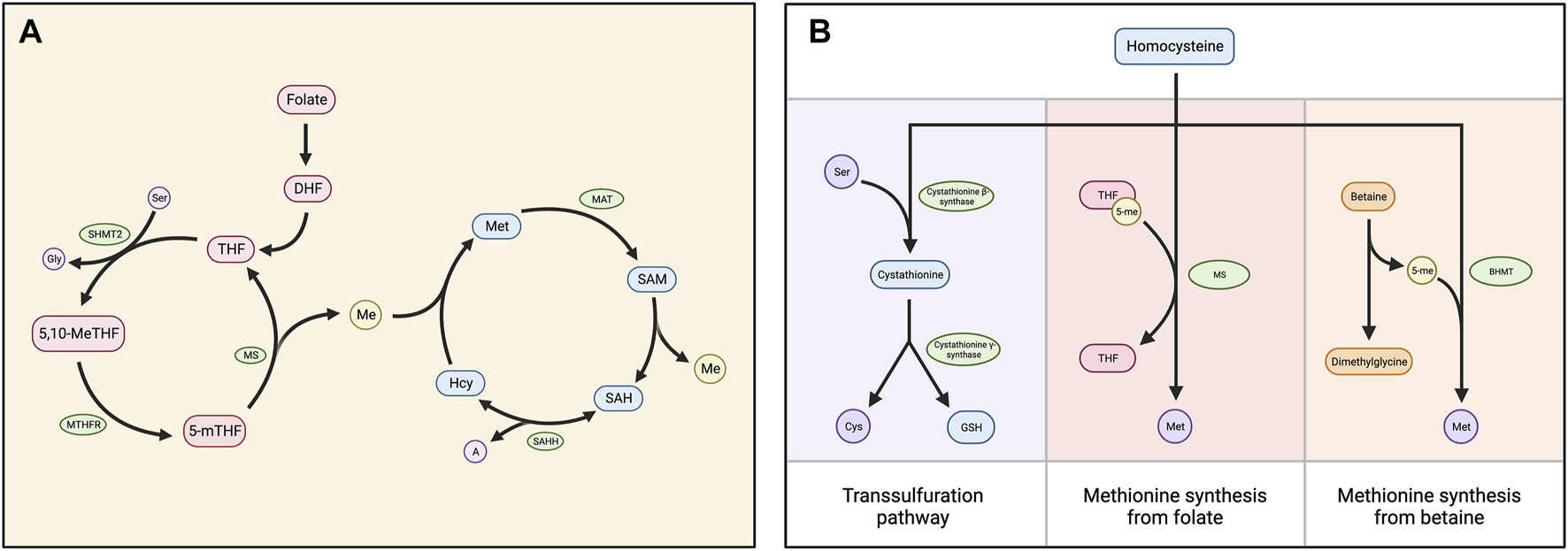
(A) Diagram depicting the folate and methionine cycles, which comprise the central mechanism of one carbon metabolism. Dietary folate is reduced to dihydrofolate (DHF) and then to tetrahydrofolate (THF), which is converted into 5,10-methylene THF (5,10-MeTHF) by serine hydroxymethyltransferase (SHMT2). 5,10-meTHF is converted to 5-methyl THF (5-mTHF) by methylenetetrahydrofolate reductase (MTHFR). Methionine synthase (MS) then transfers the methyl group from 5-mTHF to homocysteine (Hcy), generating THF and methionine (Met). Methionine adenosyltransferase (MAT) converts methionine into S-adenosylmethionine (SAM), which functions as a carbon donor in methylation reactions. This produces S-adenosylhomocysteine (SAH), which is cleaved into homocysteine (Hcy) and adenine (A) by SAH hydrolase (SAHH). (B) Chart displaying three alternatives for the processing of homocysteine. The primary mechanism is the folate cycle, in which homocysteine is converted into methionine via the MS-catalyzed transfer of a methyl group from 5-meTHF. Homocysteine can also be converted into methionine using betaine: Betaine-homocysteine methyltransferase (BHMT) catalyzes the transfer of a methyl group from betaine to homocysteine, generating methionine and dimethylglycine as a biproduct. Alternatively, the transsulfuration pathway processes homocysteine without converting it into methionine. Cystathionine β-synthase condenses homocysteine and serine (Ser) into cystathionine, which is cleaved into free cysteine and glutathione (GSH) by cystathionine β-synthase.
In addition to the reaction kinetics, feedback mechanisms regulate the efficiency of one-carbon metabolism, determining the availability of SAM and the subsequent potential for methylation reactions, called the “methylation index.” Some of these feedback mechanisms increase the methylation index. For example, glycine N-methyltransferase (GNMT), an enzyme that reduces the SAM/SAH ratio, is inhibited by 5-methyltetrahydrofolate (5-mTHF) (6, 7). Other feedbacks decrease the methylation index. For example, SAM allosterically inhibits methylenetetrahydrofolate reductase (MTHFR), while serine hydroxymethyltransferase 2 (SHMT2) sequesters 5-mTHF (6, 10). Additionally, SAH serves as a competitive inhibitor of transmethylation reactions by binding to the catalytic region of SAM-dependent methyltransferases with high affinity (5, 8, 10).
DNA methylation and demethylation pathways
DNA methylation is catalyzed by DNA methyltransferases (DNMTs), which transfer methyl groups from SAM to cytosine bases at their fifth carbon (C-5). This generates 5-methylcytosine (5-mC) that represses gene transcription (8, 11, 12) (Figure 2). 5-mC marks are most stable when they occur at CpG dinucleotides and thus are highly abundant in CpG islands (11). DNMT1 functions to maintain existing methylation patterns after DNA replication, while DNMT3a and DNMT3b induce de novo methylation (13). DNA methylation is dependent on the SAM/SAH ratio, partly because it relies on SAM availability. Additionally, due to the competitive inhibition of methyltransferases by SAH, DNMT function is dependent on the efficient hydrolysis of SAH by AHCY (8). Moreover, homocysteine re-methylation occurs primarily via the folate cycle and is necessary for the hydrolysis of SAH and regeneration of SAM. Thus, an adequate folate supply is required for efficient DNA methylation (8).
FIGURE 2
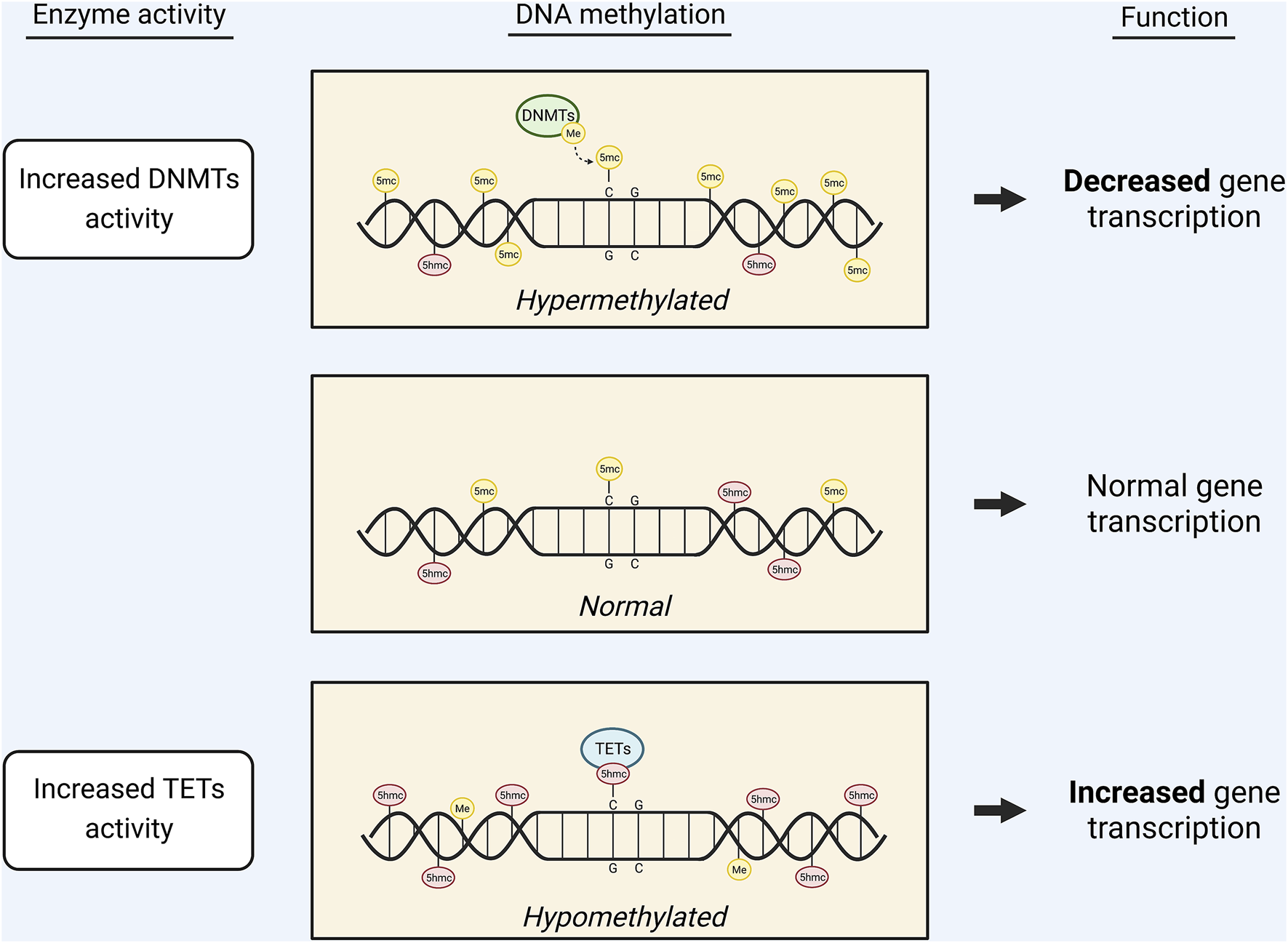
Graphic displaying the effects of DNA methylation on gene transcription. The availability of 5mc or 5hmc marks on the DNA regulates gene transcription. Increased activity of DNMTs leads to an increase in 5mc marks and, subsequently, decreased gene expression. Conversely, increased activity of TETs leads to conversion of 5mc marks into 5hmc marks, causing increased gene expression.
DNA demethylation, or the removal of 5-mC, occurs through base modifications catalyzed by ten-eleven translocation (TET) enzymes or activation-induced deaminase (AID) (14, 15). In TET-catalyzed base modification, TET 5-mc dioxygenase 1 (TET1) oxidizes 5-mC into 5-hydroxymethylcytosine (5-hmC) (8, 11) (Figure 2). TET1, TET2, and TET3 can subsequently oxidize 5-hmC into 5-formylcytosine and 5-carboxylcytosine (16). AID-catalyzed base modifications are less understood but are thought to involve AID (a member of the AID/APOBEC family of cytidine deaminase) acting on an unmodified cytosine in the vicinity of a 5-mC mark (14, 15). Both TET-catalyzed and AID-catalyzed base modifications are followed by nucleotide replacement via the base excision repair (BER) pathway (13–15).
Alcohol induces DNA hypomethylation in the liver
By altering the activity and expression of enzymes involved in one-carbon metabolism, such as methionine synthase, methionine adenosyltransferase, and betaine-homocysteine methyltransferase, alcohol consumption decreases the methylation index in the liver (16–22). As ethanol is metabolized, acetaldehyde—one of its breakdown products—irreversibly inhibits methionine synthase (MS) (16–18). Further, alcohol downregulates the mRNA expression of MTHFR and certain MAT subunits (9, 16, 19). The reduced activity of these enzymes impairs the methionine cycle’s efficiency, leading to reduced SAM availability (20). Because alcohol also downregulates the mRNA expression of betaine-homocysteine methyltransferase (BHMT), the betaine pathway is unable to generate enough SAM to compensate (19, 21, 22).
Ethanol also reduces dietary folate absorption by altering the binding and transport kinetics of the folate transport system and reducing the expression of folate transporters in the intestines and kidneys (23, 24). This folate deficiency, in addition to the inhibition of MS by acetaldehyde, contributes to the increased plasma homocysteine levels commonly seen in association with alcohol consumption (24–26). High homocysteine levels can cause the SAH hydrolysis reaction kinetics to shift in favor of the reverse reaction, leading to a build-up of hepatic SAH and a decreased SAM/SAH ratio (20, 27, 28).
Hyperactivity of the transsulfuration pathway also contributes to this reduced hepatic methylation index. This pathway is highly active in the liver under normal conditions, but it becomes even more active during chronic alcohol consumption to attenuate oxidative stress. Because the transsulfuration pathway consumes SAM without replenishing methionine, its hyperactivity leads to reduced SAM availability (29). This, in addition to the inhibition of key folate cycle and betaine pathway enzymes and increased plasma homocysteine levels due to alcohol consumption, reduces the SAM/SAH ratio, resulting in hypomethylation of genes in the liver.
The dysregulation of one-carbon metabolism and DNA hypomethylation caused by alcohol is implicated in the development of alcohol-induced liver disease (ALD) (21, 27, 29). During chronic heavy drinking, the SAM/SAH ratio decreases (20,27–29). Because the transsulfuration pathway requires SAM, the eventual depletion of SAM levels reduces the pathway’s efficacy. As a result, the liver becomes susceptible to alcohol-induced oxidative stress, which is involved in the pathogenesis of ALD (29). Folate deficiency has also been implicated in the acceleration of ALD onset, because it contributes to both SAM-deficiency and hyperhomocysteinemia (10, 27, 29). Because homocysteine is highly toxic and enhances the vulnerability of cells to oxidative injury, hyperhomocysteinemia is associated with the development of ALD (21, 26). Chronic ethanol consumption has also been shown to cause reduced expression of TET1, and the resulting decrease in DNA hydroxymethylation can contribute to hepatocyte apoptosis, ALD progression, and the development of hepatocellular carcinoma (HCC) (30–32).
Interestingly, pharmacological restoration of effective one-carbon metabolism after chronic alcohol consumption may treat alcohol-induced liver injury. Exogenous SAM administration can compensate for depleted hepatic SAM levels and effectively attenuates oxidative stress and inflammation (29, 33). In a study of human patients with alcohol-induced cirrhosis, SAM treatment was shown to improve their likelihood of survival and delay their need for liver transplantation (29). Betaine administration is also an effective treatment for ALD. It indirectly attenuates oxidative stress and apoptosis not only by increasing hepatic SAM but also by decreasing homocysteine levels (20, 29). Because the betaine pathway metabolizes homocysteine into methionine, betaine administration may be more effective than SAM administration in treating ALD (34). Additionally, betaine administration has been shown to both attenuate and reverse triacylglycerol accumulation in the liver (otherwise known as fatty liver), even despite continued ethanol consumption (27, 29).
Alcohol-induced changes in global DNA methylation in the brain
Studies of various brain regions have shown that alcohol-induced dysregulation of one-carbon metabolism can result in an increased methylation index in the brain (16, 19). In the rat and human cerebellum, alcohol consumption was found to be associated with decreased SAH levels and increased levels of adenosylhomocysteinase (AHCY), which facilitates the conversion of SAH into homocysteine (16, 19). These changes increase the SAM/SAH ratio and reduce the feedback inhibition of transmethylation reactions by SAH. In the human cerebellum, alcohol consumption was also found to be associated with increased expression of MTHFR, which promotes the formation of 5-meTHF for use in the conversion of homocysteine to methionine (16). Together, these effects of alcohol on one-carbon metabolism enzymes result in greater SAM availability and a higher potential for DNA methylation in the cerebellum.
Alcohol consumption also modulates the expression of enzymes directly involved in DNA methylation (16, 35). Interestingly, decreased TET1 expression was associated with alcohol consumption in humans, resulting in DNA hypermethylation (16). Further evidence has demonstrated that alcohol-induced hypermethylation is associated with upregulated DNMT1 and can be prevented by infusion of a DNMT inhibitor (35). It is possible that the magnitude of hypermethylation may depend on total alcohol consumption. A study of rhesus macaques found a positive correlation between CpG methylation and daily average alcohol intake (36); however, more studies are needed to confirm this finding. In summary, alcohol consumption is associated with the altered expression of enzymes central to one-carbon metabolism and DNA methylation, and these changes promote a global state of DNA hypermethylation in the brain (16, 19, 35, 36).
Gene-specific DNA methylation and altered neurobiology in AUD
Studies have identified many genes that are differentially methylated in association with alcohol consumption. The resulting changes in expression of these genes underlie a variety of phenotypes related to acute and chronic alcohol consumption, dependence, and withdrawal (4). GABAergic signaling regulates motor coordination, which is known to be disrupted by alcohol consumption. One study found decreased mRNA and protein expression of GABRD, the δ subunit of the GABAA receptor, in human AUD patients compared to controls (16). This was associated with hypermethylation of the GABRD promoter DNA. Additionally, GABRD mRNA expression was negatively correlated with number of total years the patients spent consuming alcohol. This decrease in GABRD via DNA methylation may be a component of the mechanism by which alcohol consumption impairs motor coordination (16).
Compulsive behavior during withdrawal is involved in the pathogenesis of AUD (35, 37, 38). In addition to global DNA hypermethylation and increased DNMT1 expression, Barbier et al. observed increased DNMT1 expression and downregulation of Syt2, a synaptic protein gene, in the medial prefrontal cortex (mPFC) of AUD modeled rats compared to controls (35). Infusion of DNMT inhibitor RG108 restored Syt2 expression, and lentiviral inhibition of Syt2 was found to increase aversion-resistant alcohol drinking, a compulsive behavior (35). Thus, these studies suggest that withdrawal-induced methylation of Syt2 may be implicated in the induction of compulsive behaviors and subsequent risk of AUD (35, 37, 38).
Alcohol consumption increases the activity of the endogenous opioid system (EOS), which alters reward sensitivity and cognitive control over addiction-related behaviors (39). One study identified increased prodynorphin (PDYN) expression in the postmortem dorsolateral prefrontal cortex (dlPFC) of AUD subjects. Interestingly, AUD subjects were found to have increased methylation of the C variant of 3′-UTR SNP rs2235749 (a PDYN CpG SNP known to be associated with AUD) in the dlPFC compared to control subjects, and this methylation was positively correlated with dynorphin expression. Thus, methylation of this CpG SNP may be part of the mechanism by which alcohol increases PDYN, suggesting that individuals with the C variant may be more vulnerable to developing AUD (39).
While alcohol consumption has anxiolytic effects, withdrawal from alcohol consumption results in increased anxiety. This subsequently promotes further drinking to reduce anxiety (1, 2, 3). A similar phenotype is observed in adults who first consumed alcohol during adolescence. They tend to experience higher levels of anxiety compared to alcohol-naïve adults, and this predisposes them to AUD (13). Neuropeptide Y (NPY) and brain-derived neurotrophic factor (BDNF) exert anxiolytic functions in the amygdala, and their downregulation has been observed in association with increased anxiety and alcohol consumption after adolescent alcohol exposure (13, 40–42). One study found that adolescent intermittent ethanol (AIE) exposure led to increased DNA methylation at the Npy promoter and Bdnf exon IV promoter in the adult rat amygdala in comparison to control rats, presenting a potential mechanism for the established AIE-induced downregulation of NPY and BDNF (13, 40). This methylation was associated with increased DNMT activity, higher mRNA expression of Dnmt1 and Dnmt3b, and lower mRNA expression of Gadd45g (a protein involved in demethylation) in the adult amygdala of AIE rats compared to controls (Figure 3A). Further, the AIE adult rats exhibited heightened anxiety-like and alcohol drinking behaviors compared to control rats. In support of these results, treatment with 5-azacytidine (a DNMT inhibitor) reversed the AIE-induced increases in anxiety-like behaviors and alcohol consumption and normalized the methylation status of the Npy and Bdnf promoters. Thus, adolescent alcohol exposure may lead to psychopathology associated with reduced NPY and BDNF in adulthood via DNA methylation of Npy and Bdnf (13, 40–42).
FIGURE 3
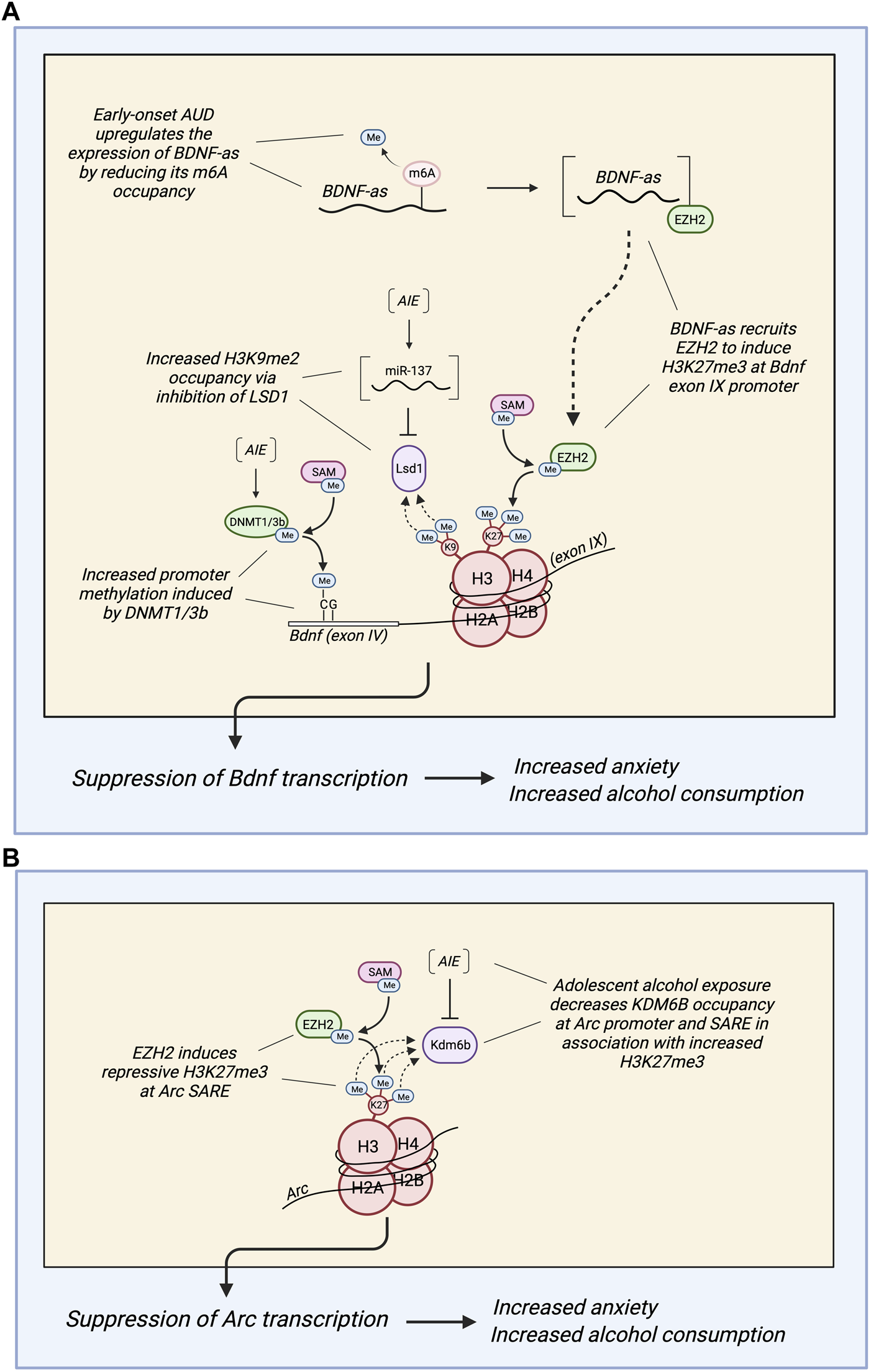
(A) Diagram depicting different epigenetic methylation mechanisms by which adolescent alcohol exposure regulates the expression of brain-derived neurotrophic factor (BDNF) in the adult amygdala of rats and humans. Adolescent intermittent ethanol (AIE) exposure produces hypermethylation of the Bdnf exon IV promoter via increased DNA methyltransferase activity (most likely due to increased expression of DNMT1/3b). AIE also increases H3K9me2 occupancy at the Bdnf exon IV promoter by upregulating miR-137, which inhibits lysine-specific histone demethylase 1 (Lsd1) (13,43). Further, AUD subjects with early age of onset are associated with reduced N6-methyladenosine (m6A) occupancy in BDNF antisense (BDNF-AS) RNA, increasing its expression. This most likely increases BDNF-AS recruitment of enhancer of zeste homolog 2 (EZH2) to induce repressive H3K27 trimethylation (H3K27me3) at the BDNF exon IX promoter and overlapping region (44). Together, these epigenetic marks induced by adolescent alcohol exposure (DNA hypermethylation, increased H3K9me2, and increased H3K27me3) suppress BDNF in the adult amygdala, leading to increased anxiety and alcohol consumption in adulthood. (B) Diagram depicting a mechanism by which adolescent alcohol exposure reduces the expression of Arc in the adult amygdala of rats and humans. AIE decreases lysine demethylase 6b (KDM6b) occupancy at the Arc promoter and SARE, which permits increased H3K27me3 at the Arc promoter and SARE site (45). In AUD subjects with early age of onset, there is an increase in EZH2 and H3K27me3 occupancy at the Arc SARE site that is associated with decreased Arc expression (44). Also, suppression of Arc expression in the adult amygdala increases anxiety and alcohol consumption in adulthood after adolescent alcohol exposure.
Alcohol-induced epigenetic modifications of genes involved in glutamatergic and GABAergic signaling have a variety of effects, such as altered synaptic plasticity and withdrawal excitotoxicity, which increase an individual’s risk of developing AUD. GPR39 is involved in the regulation of neurotransmitter release, and JAKMIP1 contributes to neurotransmitter trafficking. Hypermethylation of the GPR39 and JAKMIP1 promoter regions, in association with decreased protein expression, was observed in the NAc of rhesus macaques after chronic self-administration of alcohol. Interestingly, these effects were correlated with average daily alcohol consumption (36). Macaques with heavy levels of consumption were found to have significantly greater DNA methylation and lower protein expression of GPR39 and JAKMIP1 compared to those with low levels of consumption (36).
Alcohol consumption is also associated with altered NMDA receptor 2B (NR2B) expression that may occur via DNA methylation (46). Demethylation at the 5′ regulatory region of the NR2B gene was observed in association with increased NR2B expression in primary cortical cultured neurons after chronic intermittent ethanol treatment. The demethylation sites were located near transcription factor binding sequences for AP-1 and CRE. Interestingly, these effects persisted during withdrawal (46). This change does not reflect the established ethanol-induced global hypermethylation in the brain, but rather may occur as a neuroadaptive response to the chronic blockade of NMDA receptors by ethanol. Alternatively, other authors have suggested that because NR2B over-expression persists even when ethanol is no longer present to block NMDA receptors, the subsequent increase in excitatory neurotransmission results in withdrawal-associated excitotoxicity (47). In support of this hypothesis, it has been shown that treatment with NMDA receptor antagonists (e.g., MK-801, CGP-39551, and dizocilpine) reduces withdrawal symptoms in different models, while treatment with NMDA receptor agonists (N-methyl-D-aspartate and kainic acid) intensifies them (47).
Alcohol-induced methylation also contributes to excitatory syndrome in withdrawal via homocysteine. Because chronic ethanol consumption disrupts the blood-brain barrier, the brain is exposed to elevated homocysteine levels caused by alcohol-induced hypomethylation in the rest of the body. Because homocysteine is an excitatory amino acid, it activates metabotropic glutamate receptors (mGluRs) and NMDA receptors, causing increased glutamatergic neurotransmission. Thus, the persistent elevation of blood homocysteine levels due to ethanol contributes to the brain’s risk of hyperexcitation (25, 47). In addition to its excitotoxic effects, homocysteine is also associated with brain damage and neurodegeneration. In patients with AUD, a plasma homocysteine levels were found to correlate with reductions in hippocampal volume. Plasma homocysteine levels have also been found to correlate with brain atrophy in elderly individuals. Further, homocysteine inhibits the expression and synthesis of antioxidant enzymes and radical scavengers, which results in increased cellular apoptosis in response to oxidative stress (21, 26, 47). Thus, exposure of the brain to elevated homocysteine levels leads to overstimulation of NMDA receptors, oxidative stress, DNA damage, and endoplasmic and mitochondrial dysfunction, which may be responsible for some of the brain damage and neurodegeneration associated with AUD (21, 25, 26, 47).
The mesolimbic dopamine (DA) system has been implicated in many facets of AUD, such as craving, impulse control, seeking behavior and reward salience (48). Studies of humans, primates, and rodents have found that chronic ethanol treatment leads to reduced mesolimbic DA release (49–51). These changes in DA signaling may occur via alcohol-induced DNA methylation of key regulatory genes (52–61). Dopamine receptor D2 (D2R) (DRD2) is an inhibitory receptor that is activated in response to negative outcomes to reduce DA signaling. The downregulation of D2R results in impaired impulse control and behavioral disinhibition (52). D2R is also involved in the activation of the synaptic dopamine transporter (DAT) to attenuate high synaptic DA levels by increasing reuptake. Thus, when D2R is downregulated, the resultant decrease in DAT activation leads to hyperexcitability of DA pathways in response to rewarding stimuli. One study identified hypermethylation of DRD2 in the saliva of human AUD subjects compared to controls. DRD2 methylation was found to correlate with clinical measures of AUD severity, as well as with reward sensitivity to alcohol over appetitive cues (53). This evidence suggests that methylation of DRD2 may be implicated in the development of reward-related phenotypes associated with AUD.
Because of D2R’s role in the activation of DAT, the association between reduced D2R expression and alcohol consumption supports the possibility of a similar relationship between alcohol consumption and DAT expression (53). Interestingly, studies of DAT expression and methylation after chronic alcohol consumption have produced conflicting results. Several studies have found that chronic ethanol treatment causes decreased methylation of the DAT1 promoter, resulting in the upregulation of DAT in a variety of brain regions (54–56). However, other studies have demonstrated the opposite effect, finding that chronic alcohol consumption reduces mesolimbic DAT in association with hypermethylation of the DAT1 promoter (57–59). Some studies have also found decreased DAT expression to be associated with increased alcohol craving and voluntary alcohol consumption (60, 61).
While this evidence is still inconclusive, the possible hypermethylation of DAT1 by chronic ethanol consumption would be consistent with the decreased mesolimbic DA release associated with chronic drinking (50–52). In this condition of reduced DA release, DAT1 hypermethylation and decreased DAT expression could serve as a compensatory mechanism to maintain extracellular dopamine levels by decreasing synaptic reuptake (57, 62, 63). This would increase the sensitivity of the DA pathway to dopamine release, which could serve to potentiate the reward value of alcohol during withdrawal (60, 61). However, though this hypothesis is consistent with the known effects of alcohol on the DA system, more conclusive evidence is needed.
As a stressor, acute alcohol consumption induces glucocorticoid secretion, which increases the level of cortisol in the blood (64–66). As AUD develops, allostatic regulation of the hypothalamic pituitary adrenal (HPA) axis attenuates the accompanying chronically elevated cortisol levels, resulting in reduced cortisol responsivity to stress (1, 2, 66). The mechanism of this alcohol-induced allostatic regulation may involve modulation of glucocorticoid receptor (GR) expression. One study identified increased promoter methylation at exon 1H of NR3C1, which encodes GR, in postmortem brain tissue of human AUD subjects compared to controls (64). This hypermethylation was associated with reduced mRNA and protein GR expression in the hippocampus, amygdala, striatum, and PFC (Brodmann Area 10), as well as altered expression of several other stress-responsive genes in the PFC (64). This evidence suggests that the body’s allostatic mechanism for tolerance to chronic alcohol-induced glucocorticoid secretion involves GR downregulation via methylation of NR3C1. Additionally, because NR3C1 also serves to enhance DRD2 expression, this evidence is consistent with the previously discussed downregulation of DRD2 in AUD (53, 67).
Histone methylation
Methylation of DNA and histones is often locally coordinated to exert concordant effects on gene transcription (68–70). Like DNA methylation, histone methylation is dependent on SAM (5, 7). However, histone methylation does not universally increase or decrease based on the methylation index. Rather, because histone methylation marks can be repressive or activating, they are often deposited in such a way that reinforces the transcriptional effects of a particular gene’s DNA methylation status.
Methylation can occur on lysine residues in histones H3 and H4, and each lysine can be mono-, di-, or tri-methylated (71–73). Lysine methylation is regulated by methyltransferases (KMTs or “writers”) and demethylases (KDMs or “erasers”) (74). These enzymes are highly specific to individual lysine residues and particular degrees of methylation (72–74). Effector proteins (or “readers”) are responsible for recognizing a methylated lysine site and exerting the corresponding effect on gene expression (75).
The primary KMT class is SET-domain-containing enzymes (74, 75). Two of these, G9a and GLP, form a functional complex through their SET domains and can induce methylation at H3K9 and H3K27 (76). The G9a–GLP complex is particularly associated with the induction of H3K9me2, which functions to downregulate gene expression (76). Other SET family KMTs are MLL1 and MLL2, both of which induce methylation at H3K4 (56). Additionally, enhancer of zeste homolog 2 (EZH2) induces repressive trimethylation of H3K27 (H3K27me3) (42).
One important KDM is lysine-specific histone demethylase 1 (LSD1 or KDM1A/B) (40, 71, 72). LSD1 reverses H3K4me1/2 and H3K9me1/2 via an amine oxidase reaction using flavin as a cofactor (40, 71). Several other KDMs are Jumonji C domain-containing (JmjC) demethylases, or JHDMs (40, 72, 74). JHDM1 demethylates H3K36, JHDM2A-D and JHDM3 demethylate H3K9 and H3K36, and JARID1C (JmjC AT-rich interactive domain 1C enzyme) demethylates H3K4me2/3 (40, 77).
Histone lysine methylations have differential effects on gene expression (Figure 4): They can be found at both active and inactive open reading frames depending on the specific lysine residue and degree of methylation (73, 74). Activating marks typically include H3K4me3, and H3K36me3, whereas H3K27me2/3, H3K9me2/3, and H4K20me3 are typically repressive (72, 75, 76). However, certain mono-methylation marks—such as H3K9me, H3K36me, and H3K4me—can cause activation or repression of gene expression (77, 78) (Figure 4).
FIGURE 4
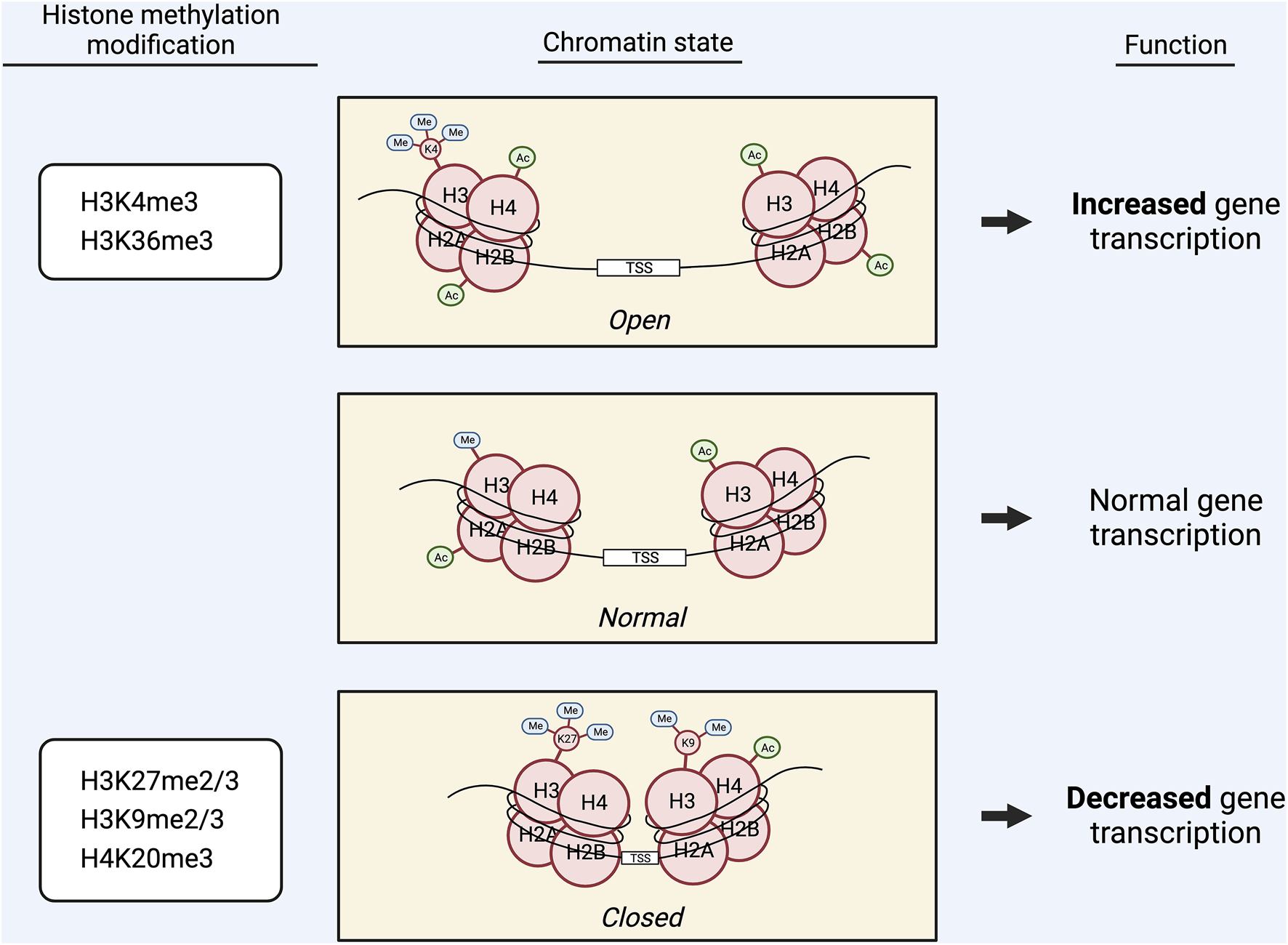
Graphic displaying the effects of methylation of different lysine (K) residues in histones H3 and H4 on chromatin state and gene transcription. Activating marks, including H3K4me3 and H3K36me3, induce a more open chromatin state, which promotes gene transcription. Repressive marks, including H3K27me2/3, H3K9me2/3, and H4K20me3, induce a more closed chromatin state, which decreases gene transcription. Alternatively, certain mono-methylation marks, such as H3K9me, H3K36me, and H3K4me, are capable of exerting both activating and repressive effects on gene transcription.
Alcohol alters histone methylation in the liver
Though chronic alcohol consumption decreases the methylation index in the liver, this does not result in a global decrease in histone methylation as with DNA methylation. Instead, chronic alcohol consumption results in histone methylation changes that reinforce the transcription-promoting effects of global DNA hypomethylation: increased activating marks and decreased repressive marks (79, 80). This was demonstrated in hepatocytes treated with ethanol, which showed reduced repressive mark, H3K9me2, and increased active mark, H3K4me2, in association with a two-fold increase in about 35% of the genes expressed in the liver (79, 80). This evidence suggests that alcohol’s effect on histone methylation in the liver serves to contribute to a global state of increased gene transcription, in concordance with alcohol-induced DNA hypomethylation (79, 80).
Alcohol-induced changes in histone methylation, as with DNA methylation, contribute to the pathophysiology of ALD. Alcohol has been found to increase the hepatic activity of MLL1, a SET-domain-containing KMT that induces H3K4me2/3 in hepatic stellate cells. This results in accelerated trans-differentiation of these cells, leading to increased net deposition of fibril-forming extracellular matrix. This process eventually leads to fibrosis, a common feature of ALD (81).
Thus, alcohol-induced changes in histone methylation in the liver mirror those of DNA methylation. Though histone methylation is not globally reduced in the liver, it is still altered in such a way that reinforces the transcription-promoting effects of DNA hypomethylation: Repressive marks are decreased, and activating marks are increased. Further, alcohol’s effects on histone methylation contribute to the development of ALD, in combination with its effects on DNA methylation (79–81).
Alcohol induces specific changes in histone methylation in the brain
Alcohol consumption induces a variety of changes in histone methylation that contribute to phenotypes associated with the pathophysiology of AUD. As previously discussed, the EOS of AUD individuals is characterized by increased PDYN because of methylation at particular SNPs in the PDYN promoter, which promotes anxiogenesis (39). Another study found that acute alcohol caused increased DYN and NOC in association with increased H3K9Ac and decreased H3K27me3 at the Pdyn and Pnoc promoters in the amygdala of treated rats compared to controls. This may reflect the role of acute alcohol as both an anxiolytic and an acute stressor (82–84).
As previously discussed, DNA methylation is implicated in the induction of anxiety in adult rats that were exposed to alcohol during adolescence (Figure 3A) (13, 40). Studies have demonstrated similar effects of AIE on histone methylation (43). Decreased Arc eRNA expression was observed in the amygdala of adult AIE rats compared to controls. This was associated with increased anxiety-like behaviors as well as decreased KDM6B occupancy and increased H3K27me3 occupancy at the Arc synaptic activity response element (SARE) site and promoter. Interestingly, knockdown of KDM6B via siRNA infusion induced the same effects in alcohol-naïve adults: increased anxiety-like behaviors, decreased Arc eRNA and mRNA expression, decreased KDM6B occupancy, and increased H3K27me3 occupancy at Arc regulatory sites (43). This evidence suggests that AIE leads to increased H3K27me3 at the Arc promoter by downregulating the activity of KDM6B, causing decreased Arc expression and increased anxiety-like behaviors in adulthood (Figure 3B). A subsequent study confirmed this epigenetic remodeling using targeted epigenomic editing (85). Infusion of dCas9-P300 into the CeA increased H3K27Ac at Arc SARE, increased Arc eRNA and mRNA expression, and ameliorated the AIE-induced anxiety and excessive alcohol intake in adult rats. Further, infusion of dCas9-KRAB into control rats led to increased repressive H3K27me3 at Arc SARE, decreased Arc eRNA and mRNA expression, and the development of anxiety and alcohol drinking behaviors (85). Thus, these findings causally link epigenetic modifications of Arc at H3K27 with the induction of anxiety and alcohol consumption phenotypes in adulthood as a result of AIE (85).
In addition to DNA hypermethylation, histone methylation changes also contribute to the AIE-induced downregulation of Bdnf. A study found decreased Bdnf expression in the amygdala of AIE adult rats compared to controls in association with increased H3K9me2 occupancy at the Bdnf exon IV promoter. This was also associated with decreased Lsd1 and Lsd1+8a mRNA expression, decreased LSD1 protein expression, and decreased LSD1 binding to the Bdnf exon IV promoter, as well as increased anxiety-like behaviors and voluntary ethanol consumption (40). A subsequent study demonstrated increased expression of miR-137, a microRNA that targets Lsd1, in the amygdala of AIE adult rats compared to controls (40, 41). Exposure of AIE adult rats to acute ethanol normalized the AIE-induced phenotypes to those of control rats: the heightened anxiety-like behaviors were attenuated, the decreased Lsd1+8a mRNA expression was normalized, and the increased H3K9me2 occupancy at the Bdnf exon IV promoter was normalized (40). Interestingly, infusion of a miR-137-specific antagomir into the CeA also reversed these AIE-induced phenotypes: anxiety-like behaviors were attenuated, voluntary ethanol consumption was normalized, the decreased Lsd1 and Lsd1+8a mRNA expression was rescued, the decreased LSD1 binding to the Bdnf IV promoter was restored, the increased H3K9me2 occupancy at the Bdnf IV promoter was normalized, and the decreased expression of Bdnf was rescued. Notably, co-infusion of Lsd1 siRNA prevented the normalization of these phenotypes, indicating that LSD1 is an essential component of this mechanism (41). In summary, these results suggest that AIE increases miR-137 in the adult amygdala, which allows increased H3K9me2 occupancy at the Bdnf exon IV promoter via the inhibition of Lsd1. This modulation, in addition to the AIE-induced DNA hypermethylation of the Bdnf exon IV promoter, results in reduced BDNF, which contributes to the development of anxiety-like behaviors and increased voluntary alcohol consumption in adulthood (13, 40, 41) (Figure 3A). In addition to its role in AIE-induced adult anxiety, NPY is also involved in the anxiolytic effects of acute ethanol (13, 86). Decreased H3K9me2 and G9a occupancy at the regulatory region of Npy has been observed in rats treated with acute ethanol in association with increased NPY expression and decreased anxiety-like behaviors (86). Thus, altered histone methylation of NPY appears to be involved in anxiolysis due to acute ethanol.
As previously discussed, increased NR2B expression is implicated in multiple phenotypes of AUD. In addition to DNA hypomethylation of the NR2B promoter, histone methylation changes also appear to contribute to the alcohol-induced upregulation of NR2B. One study demonstrated that chronic intermittent ethanol exposure causes increased H3K9ac and decreased H3K9me2/3 occupancy at the NR2B promoter in a cortical neuronal cell culture, resulting in increased NR2B expression. This was observed in association with decreased expression of multiple KMTs, including G9a (87).
Alcohol may also affect GABAergic signaling by modulating the expression of GABA-Aɑ5, a subunit of GABAA. Studies have shown that GABAA receptor activity is associated with a variety of alcohol tolerance, dependence, and withdrawal phenotypes, and GABA-Aɑ5 has been implicated in alcohol-induced memory loss and cognitive impairment (16, 88–90). Increased GABA-Aɑ5 expression has been observed in association with increased H3K4me3 and H3K9ac occupancy in the PFC of rats treated with chronic ethanol compared to controls (88). Interestingly, increased GABA-Aɑ5 expression and H3K4me3 occupancy has also been observed in rats with a genetic background of AUD, and this was associated with increased vulnerability to alcohol addiction. Because of this association, alcohol-induced upregulation of GABA-Aɑ5 via histone methylation may increase the risk that an individual and their offspring will develop AUD (88–90).
One study identified increased expression of four genes related to ethanol tolerance (HSP104, PRO1, TPS1, and SOD1) in ethanol-tolerant strains of S. cerevisiae in association with enhanced H3K4me3 occupancy at the promoter of each (91). After passage through a stress-free medium treatment, the expression of these genes and H3K4me3 occupancy was significantly decreased. This evidence suggests that H3K4me3 is involved in mediating the ethanol tolerance of these strains (91). Another study also found evidence supporting the role of histone methylation in ethanol tolerance (92). Individual knockout of four JHDMs (NO66, KDM3, KDM5B, and HSPBAP1) in drosophila resulted in differential changes in ethanol tolerance and sensitivity to ethanol-induced sedation (92, 93). Thus, these studies suggest that histone demethylases may be required for normal ethanol tolerance and ethanol-induced sedation (91–93).
RNA methylation
While the involvement of epigenetic mechanisms in the pathophysiology of AUD has been extensively characterized (3,4), far less is known about the role of epitranscriptomics (RNA epigenetics). Broadly, epitranscriptomics describes molecular modifications of RNA, including messenger RNA, transfer RNA, ribosomal RNA, and non-coding RNAs, that exert functional changes (94). There are many known RNA modification types, but the most widely studied involve the methylation of guanine, cytosine, and adenine. As compared to those of guanine and cytosine, adenine modifications have been the most extensively investigated. Adenine can be methylated in diverse ways, but the most prevent is the m6A modification, which accounts for approximately 50% of all methylated ribonucleotides (94). M6A modifications are reversible and have been shown to control splicing, stability, and translation of the transcriptome (94, 95). Strikingly, the mechanism of m6A methylation also involves the usage of SAM, which is part of one-carbon metabolism. This connection suggests m6A methylation also plays an important role in the fine tuning of gene expression in AUD, and understanding how cells manage their one-carbon metabolism and output of SAM for the all methylation mechanisms discussed herein are perhaps indispensable to developing treatments for AUD. Furthermore, the m6A methylation could be under the control of epigenetic mechanisms, and such a feature would provide a mechanistic bridge between gene regulation and gene expression (95). Thus, m6A modified RNAs, especially mRNAs, present an enticing avenue for exploration into the pathophysiology of AUD.
M6A methylation is conferred to RNAs co-transcriptionally by the METTL3–METTL14 methyltransferase (writer) complex, and its associated ancillary proteins. METTL3 (methyltransferase-like 3) is the catalytically active subunit, while METTL14 (methyltransferase-like 14) functions as a holding scaffold that facilitates RNA binding to the complex (96–98). Of the ancillary writers, Wilm’s tumor 1-associating protein (WTAP) is the most notable, and it is known to recruit the METTL3-14 complex to nuclear speckles (97, 98) (Figure 5). Although some m6A sites are WTAP-dependent and others are WTAP-independent, knockdown of WTAP is known to drastically reduce m6A mRNA levels (94, 98).
FIGURE 5
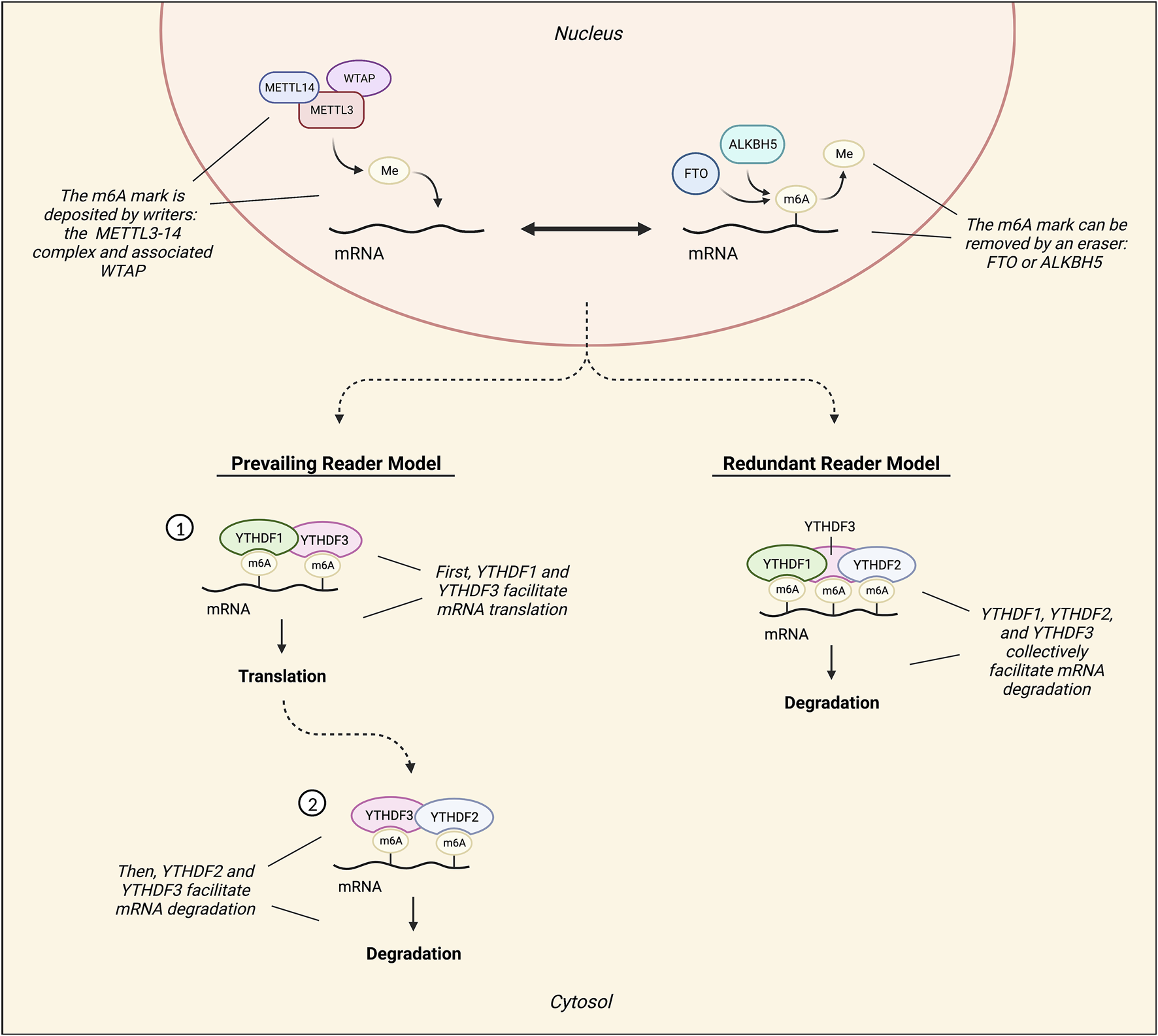
Diagram illustrating the mechanisms involved in RNA m6A methylation. The METTL3-14 methyltransferase (writer) complex, which can be assisted by ancillary writers such as Wilm’s tumor 1-associating protein (WTAP), confers m6A methylation marks. Conversely, demethylases ALKBH5 and FTO remove m6A marks. Readers are responsible for processing and defining the fate of m6A RNAs. The most characterized readers are YTHDF1, YTHDF2, and YTHDF3, and there are two contrasting models of their functionality: the prevailing model and the redundant model. According to the prevailing model, an m6A mark is first read by YTHDF3 with the assistance of YTHDF1. YTHDF1 then recruits translation factors to promote the translation of the marked mRNA. Following this, the transcript is inevitably read by YTHDF2 under facilitation by YTHDF3 to promote its decay. Conversely, the redundant reader model suggests that instead of exerting independent, disparate functions on marked transcripts, these readers interact with marked mRNA transcripts collectively to promote its degradation.
Two m6A demethylases (erasers) have been characterized thus far, ALKBH5 and FTO (Figure 5). ALKBH5 directly removes the methyl group from an m6A mark without oxidizing (97, 98). FTO, on the other hand, reverses both m6A and m6Am via oxidation, but has a higher catalytic activity for m6Am (96, 97). FTO is also known as the “obesity risk gene” because its dysregulation, particularly via common SNP variants, has been associated with obesity (99).
M6A readers process and define the fate of m6A RNAs. The most characterized group of m6A readers is the YT521-B homology domain family (YTHDF), which is comprised of YTHDF1, YTHDF2, and YTHDF3 (96–98). According to the prevailing m6A reader model, YTHDF1 functions to promote mRNA translation by recruiting various translation factors, such as eIF3, to marked mRNAs (95, 98). In contrast, YTHDF2 facilitates the localization of marked mRNA transcripts to processing bodies for degradation (94, 97, 98) (Figure 5). However, certain stress conditions can trigger the localization of marked transcripts to stress granules instead, where they are temporarily protected from degradation (94). Additionally, YTHDF2 may also function to preserve 5′UTR m6A marks on stress-induced transcripts by preventing FTO-mediated demethylation in the nucleus (98). Lastly, YTHDF3 does not exert independent effects, but rather appears to assist in the functions of both YTHDF1 and YTHDF2 through temporal patterns of interaction, first facilitating mRNA translation with YTHDF1 and then committing mRNAs for degradation with YTHDF2 (100) (Figure 5).
Conversely, an alternative model of m6A reader function has been suggested: the redundant reader model (101). This model postulates that the readers do not confer biologically significant and disparate effects independently, but rather, function cooperatively to promote m6A RNA decay (94, 101, 102) (Figure 5). In mouse embryonic stem cells, only the simultaneous knockout of all YTHDFs decreased the degradation rate of m6A mRNAs, while knockout of individual YTHDFs did not result in significantly decreased degradation rates (102). Moreover, additional X-ray crystallography and molecular dynamics data have demonstrated that the YTHDFs all interact with m6A mRNAs in the same way (103). In light of the discrepancies between this model and the prevailing m6A reader model, it appears particularly important to study all three YTHDFs collectively whenever possible.
Implication of m6A in liver and kidney pathology
While studies are lacking on the role of m6A methylation in ALD specifically, many studies have found that m6A regulation is involved in adipogenesis, a hallmark symptom of ALD. Decreased m6A via oxidation by FTO has been shown to cause increased adipogenesis (104, 105). Increased FTO mRNA and protein levels have been observed in the liver in patients with non-alcoholic fatty liver disease (104). Further, Hu et al. found that FTO is activated by glucocorticoid receptors to erase m6A marks on the mRNA of lipogenic genes (106). Conversely, because it deposits m6A, METTL3 activity has been found to negatively correlate with adipogenesis. YTHDF2 activity also negatively correlates with adipogenesis, as it can induce the degradation of m6A-marked lipogenic transcripts (104).
In addition to liver disease, kidney inflammation is a common consequence of chronic alcohol consumption. Yu et al. identified a potential mechanism for this inflammation via m6A methylation. In a mouse model of alcohol-induced kidney injury, alcohol was found to cause increased DNA methylation of FTO in the kidneys, subsequently reducing its expression. This was confirmed via inhibition of DNMTs, which attenuated alcohol-induced kidney injury and reversed the downregulation of FTO. Alcohol was also found to cause increased YTHDF2 mRNA and protein levels in the kidneys. Based on this evidence, Yu et al. proposed that alcohol-induced downregulation of FTO increases m6A methylation of peroxisome proliferator-activated receptor alpha (PPAR-ɑ). These marks are then read by YTHDF2, destabilizing and promoting degradation of PPAR-ɑ mRNA, leading to inflammation (107). This data further suggests that knocking down METTL3 may also confer a protective effect against kidney inflammation induced by chronic alcohol consumption. However, differential regulation of the m6A proteins in various tissues is incredibly complex, so these hypothesized mechanisms require further investigation.
Relationship between m6A and alcohol in the brain
M6A methylation appears to be another mechanism by which alcohol consumption alters BDNF expression in the amygdala. Increased levels of BDNF antisense (BDNF-AS), a long non-coding RNA (lnc-RNA) that regulates BDNF expression, have been found in the postmortem amygdala of individuals with AUD who began drinking during adolescence compared to those with late-onset AUD and control subjects (42). Interestingly, there was a significant reduction in BDNF-AS RNA methylation at m6A sites in adolescent-onset AUD but not in late-onset AUD. In light of the ability of m6A methylation to promote degradation and alter the recruitment of RNA-binding proteins, the authors suggest that the decrease in m6A methylation of BDNF-AS may play a causal role in its upregulation in adolescent-onset AUD (42). BDNF-AS most likely recruits EZH2 to induce H3K27me3 at the BDNF exon IX promoter and overlapping region, which inhibits BDNF expression. Because BDNF activates ARC transcription, ARC expression may also be indirectly downregulated by BDNF-AS. The decreased expression of both genes as a result of BDNF-AS upregulation is associated with increased anxiety, alcohol consumption, and risk of AUD (40, 42). This epigenetic regulation of BDNF and ARC expression in early-onset AUD is consistent with other AIE-induced modifications found in previously discussed studies in the rat model: Increased H3K27me3 occupancy at the Arc SARE and promoter sites, DNA hypermethylation of the Bdnf exon IV promoter, and increased H3K9 occupancy at the Bdnf exon IV promoter (13, 43, 41). These epigenetic marks all contribute to the downregulation of BDNF and Arc, which has been shown to result in increased anxiety and alcohol consumption (13, 42, 41) (Figure 3B).
Studies assessing the effects of the rs9939609a FTO variant on an individual’s risk of AUD have produced mixed results. Some have found rs9939609a to be a risk factor for alcohol dependence (AD). An analysis of two large-scale Caucasian sample groups yielded significant associations between AD and several FTO SNPs, including the rs9939609a variant (108). Similarly, an association between AD and rs8050136a, which is in complete linkage disequilibrium with rs9939609a, was found in a mixed-ethnicities sample (109). Contrarily, other studies have found rs9939609a to be a protective factor against AD. In a Caucasian sample, rs9939609a was associated with reduced alcohol consumption (110). Likewise, rs9939609a was found to be associated with less frequent drinking, increased alcohol consumed per drinking instance, and reduced overall alcohol consumption in another analysis of a Caucasian sample (111). However, the authors of this study did not control for other variables, such as BMI or age, when comparing AD subjects to controls (109). Another study found no association, whether positive or negative, between rs9939609a and alcohol intake (112).
Interestingly, alcohol consumption and AD risk may moderate the effects of FTO obesity-risk SNPs on BMI. Genetic obesity risk based on FTO SNPs has been shown to have a larger effect on BMI in infrequent drinkers: about twice as large in those who never drank alcohol compared to every-day drinkers. This effect decreased with increased alcohol consumption frequency, and there was an overall inverse correlation between alcohol intake frequency and BMI (113). Alternatively, another study found that males with a high risk of AD had overall significantly higher BMI during adolescence compared to low-risk males. Further, high-risk males who were homozygous for the obesity-associated allele of an FTO SNP had higher BMI during late adolescence compared to those who carried the minor allele (114). Although studies have produced mixed results regarding the effects of obesity-risk FTO alleles on risk of AUD, alcohol consumption and AD risk do appear to moderate the effects of obesity-risk FTO alleles on BMI. More studies are needed to discern whether the relationship between alcohol and FTO SNPs is causal, as well as whether there are bidirectional effects.
Avenues for research on alcohol and m6A
Studies regarding the relationship between alcohol and m6A methylation are limited. However, existing literature has identified m6A methylation involvement in particular gene networks that are central to AUD pathophysiology. These networks pose avenues for research into the potential relationships between alcohol consumption and m6A methylation.
Several studies have demonstrated the involvement of FTO-mediated m6A methylation in the regulation of reward pathways, including the dopamine system (115–120). Overexpression of FTO in rats was found to cause increased expression of glutamate receptor subunit 1 (Nmdar1) via decreased m6A methylation. The resulting oxidative stress and Ca2+ influx led to apoptosis of dopaminergic neurons (116). In another study, inactivation of FTO in mice resulted in impaired D2/3R autoinhibition of midbrain dopaminergic neurons in association with increased m6A levels in the midbrain and striatum (115). Additionally, the rs9939609a variant of FTO has been associated with impaired D2/3R signaling and increased reward sensitivity (117, 119, 120). In light of the previously discussed dopaminergic effects of alcohol, particularly the downregulation of D2R signaling and increased reward sensitivity seen in individuals with AUD, further research should explore the effects of alcohol on FTO-mediated m6A methylation in the dopaminergic system (53).
Endogenous glucocorticoids involved in the acute stress response have been shown to modulate the expression of FTO, ALKBH5, and METTL3 via glucocorticoid response elements in their 5′ upstream regions, causing altered m6A levels across multiple brain regions in both humans and rodents. Interestingly, knockout of METTL3 and FTO has been shown to result in abnormal stress-coping behaviors, suggesting that the modulation of m6A by glucocorticoids plays a functional role in the stress response (121). As a stressor, acute alcohol consumption stimulates glucocorticoid secretion, causing increased blood cortisol levels (64, 65, 66). Additionally, high-risk social drinkers tend to have enhanced cortisol responsivity to stress, while AUD individuals have reduced cortisol responsivity to stress because of allostatic regulation (64, 66, 120). Considering the effect of glucocorticoids on m6A levels, it would be interesting to investigate how m6A levels are differentially affected by acute and chronic alcohol consumption, risk factors for AUD, and allostatic regulation. Further, elucidating more of the downstream effects of stress-induced modulation of m6A in the PFC and amygdala, in addition to the abnormal stress-coping behaviors observed in response to METTL3- and FTO-knockout, could provide valuable insights into their potential role in alcohol-related pathophysiology.
Conclusion and future directions
This review has presented evidence of global and gene-specific epigenetic methylation modifications in the liver and brain produced by alcohol consumption. Though DNA, histone, and RNA methylation all rely on one-carbon metabolism for the generation of methyl groups, they are differentially affected by alcohol.
DNA methylation is directly influenced by the methylation index, which relies on the efficiency of one-carbon metabolism. In the liver, alcohol-induced dysregulation of one-carbon metabolism produces a decreased methylation index. The resulting DNA hypomethylation leads to changes in gene expression that underlie elements of the pathophysiology of ALD (9, 19, 22). Conversely, chronic alcohol consumption may cause an increased methylation index in the brain, leading to global DNA hypermethylation. The subsequent changes in gene expression contribute to several components of the brain pathophysiology of AUD that promote continued alcohol consumption, including withdrawal-induced anxiety, reduced behavioral inhibition, and increased reward sensitivity (16, 19, 35).
In contrast, histone methylation does not globally increase or decrease based on the methylation index. Rather, alcohol-induced histone methylation modifications often reinforce the transcriptional effects of alcohol-induced DNA methylation. In the liver, this occurs globally. Chronic alcohol consumption leads to an increased abundance of activating histone methylation marks (79, 80). In the brain, most effects of alcohol on histone methylation are gene-specific, resulting in changes in gene expression that are consistent with alcohol’s effects on DNA methylation and contribute to the pathophysiology of AUD (82–92).
Compared to DNA and histone methylation, far less is known about RNA m6A methylation and its relationship with alcohol consumption. Studies should explore whether alcohol modulates the expression and activity of RNA modifiers and the prevalence of m6A methylation in any global or region-specific ways. Elucidating these general effects will better inform subsequent research regarding the effects of alcohol on gene-specific m6A methylation. Several interesting avenues for such studies have already emerged from existing literature (95). Other RNA methylation marks (such as m5C, m7G, and m1A) should also be studied to develop a more complete picture of the links between alcohol consumption and RNA methylation. Insight into these relationships and their mechanisms will develop a new dimension of the field’s understanding of AUD pathophysiology, which will hopefully inform novel approaches to the treatment and prevention of AUD.
Statements
Author contributions
SP conceived the idea and discussed with TC, KK, and EM. TC and SP prepared figures and edited by all authors. All authors wrote, edited and approved the final version of the manuscript.
Funding
SP is supported by National Institute on Alcohol Abuse and Alcoholism grants (UO1AA019971, U24AA024605, R01AA025035, and P50AA022538) and the Department of Veterans Affairs (Merit Grant, I01 BX004517 and Senior Research Career Scientist Award, IK6BX006030). EM is supported by a post-doctoral fellowship within NIAAA alcohol research training grant (T32 AA026577).
Acknowledgments
Figures were prepared using BioRender.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author disclaimer
The content and discussion presented in this review is solely responsibility of the authors and does not represent the official views of the National Institutes of Health or U.S. Department of Veterans Affairs.
Glossary
- 5-mC
5-methylcytosine
- AD
alcohol dependence
- AHCY
SAH hydrolase
- AIE
adolescent intermittent ethanol
- ALD
alcohol-induced liver disease
- ALKBH5
AlkB homolog 5
- ARC
activity-regulated cytoskeleton associated protein
- AUD
alcohol use disorder
- BDNF
brain-derived neurotrophic factor
- CRE
cAMP response element
- DA
dopamine
- dlPFC
dorsolateral prefrontal cortex
- DNMT
DNA methyltransferase
- DYN
dynorphin
- EOS
endogenous opioid system
- eRNA
enhancer RNA
- EZH2
enhancer of zeste homolog 2
- FTO
fat mass and obesity-associated protein
- GABA
gamma-aminobutyric acid
- GR
glucocorticoid receptor
- KDM
lysine demethylase
- KMT
lysine methyltransferase
- M6A
N6-methyladenosine
- METTL14
methyltransferase-like 14
- METTL3
methyltransferase-like 3
- mPFC
medial prefrontal cortex
- MS
methionine synthase
- NAc
nucleus accumbens
- NMDA
N-methyl-D-aspartate
- NOC
nociceptin
- NPY
neuropeptide Y
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- SARE
synaptic activity response element
- SNP
single nucleotide polymorphism
- WTAP
Wilm’s tumor 1-associating protein
- YTHDF
YT521-B homology domain family
References
1.
Becker HC . Effects of alcohol dependence and withdrawal on stress responsiveness and alcohol consumption. Alcohol Res (2012) 34(4):448–58.
2.
Koob GF Le Moal M . Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology (2001) 24(2):97–129. 10.1016/S0893-133X(00)00195-0
3.
Pandey SC Kyzar EJ Zhang H . Epigenetic basis of the dark side of alcohol addiction. Neuropharmacol (2017) 122:74–84. 10.1016/j.neuropharm.2017.02.002
4.
Bohnsack JP Pandey SC . Histone modifications, DNA methylation, and the epigenetic code of alcohol use disorder. Int Rev Neurobiol (2021) 156:1–62. 10.1016/bs.irn.2020.08.005
5.
Mentch SJ Locasale JW . One carbon metabolism and epigenetics: Understanding the specificity. Ann NY Acad Sci (2016) 1363(1):91–8. 10.1111/nyas.12956
6.
Ducker GS Rabinowitz JD . One-carbon metabolism in health and disease. Cell Metab (2017) 25(1):27–42. 10.1016/j.cmet.2016.08.009
7.
Serefidou M Venkatasubramani AV Imhof A . The impact of one carbon metabolism on histone methylation. Front Genet (2019) 10:764. 10.3389/fgene.2019.00764
8.
Friso S Udali S De Santis D Choi S-W . One-carbon metabolism and epigenetics. Mol Aspects Med (2017) 54(1):28–36. 10.1016/j.mam.2016.11.007
9.
Lu SC Mato JM . Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol (2005) 35(3):227–34. 10.1016/j.alcohol.2005.03.011
10.
Schalinske KL Nieman KM . Disruption of methyl group metabolism by ethanol. Nutr Rev (2005) 63(11):387–91. 10.1111/j.1753-4887.2005.tb00375.x
11.
Anderson OS Sant KE Dolinoy DC . Nutrition and epigenetics: An interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem (2012) 23(8):853–9. 10.1016/j.jnutbio.2012.03.003
12.
Holliday R Grigg GW . DNA methylation and mutation. Mutat Res Fundam Mol Mech Mutagenesis (1993) 285(1):61–7. 10.1016/0027-5107(93)90052-h
13.
Sakharkar AJ Kyzar EJ Gavin DP Zhang H Chen Y Krishnan HR et al Altered amygdala DNA methylation mechanisms after adolescent alcohol exposure contribute to adult anxiety and alcohol drinking. Neuropharmacology (2019) 157:107679. 10.1016/j.neuropharm.2019.107679
14.
Bochtler M Kolano A Xu G-L . DNA demethylation pathways: Additional players and regulators. Bioessays (2017) 39(1):1–13. 10.1002/bies.201600178
15.
Guo JU Su Y Zhong C Ming G Song H . Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell (2011) 145(3):423–34. 10.1016/j.cell.2011.03.022
16.
Gatta E Auta J Gavin DP Bhaumik DK Grayson DR Pandey SC et al Emerging role of one-carbon metabolism and DNA methylation enrichment on δ-containing GABAA receptor expression in the cerebellum of subjects with alcohol use disorders (AUD). Int J Neuropsychopharmacol (2017) 20(12):1013–26. 10.1093/ijnp/pyx075
17.
Kenyon SH Nicolaou A Gibbons WA . The effect of ethanol and its metabolites upon methionine synthase activity in vitro. Alcohol (1998) 15(4):305–9. 10.1016/s0741-8329(97)00134-1
18.
Barak AJ Beckenhauer HC . The influence of ethanol on hepatic transmethylation. Alcohol (1988) 23(1):73–7.
19.
Auta J Zhang H Pandey SC Guidotti A . Chronic alcohol exposure differentially alters one-carbon metabolism in rat liver and brain. Alcohol Clin Exp Res (2017) 41(6):1105–11. 10.1111/acer.13382
20.
Kharbanda KK . Alcoholic liver disease and methionine metabolism. Semin Liver Dis (2009) 29(2):155–65. 10.1055/s-0029-1214371
21.
Barak AJ Beckenhauer HC Kharbanda KK Tuma DJ . Chronic ethanol consumption increases homocysteine accumulation in hepatocytes. Alcohol (2001) 25(2):77–81. 10.1016/s0741-8329(01)00168-9
22.
Tsukamoto H Lu SC . Current concepts in the pathogenesis of alcoholic liver injury. FASEB J (2001) 15(8):1335–49. 10.1096/fj.00-0650rev
23.
Hamid A Wani NA Kaur J . New perspectives on folate transport in relation to alcoholism-induced folate malabsorption – association with epigenome stability and cancer development. FEBS J (2009) 276(8):2175–91. 10.1111/j.1742-4658.2009.06959.x
24.
Blasco C Caballeria J Deulofeu R Lligona A Pares A Lluis JM et al Prevalence and mechanisms of hyperhomocysteinemia in chronic alcoholics. Alcohol Clin Exp Res (2005) 29(6):1044–8. 10.1097/01.alc.0000169265.36440.ee
25.
Bleich S Degner D Wiltfang J Maler JM Niedmann P Cohrs S et al Elevated homocysteine levels in alcohol withdrawal. Alcohol Alcohol (2000) 35(4):351–4. 10.1093/alcalc/35.4.351
26.
Bleich S Bandelow B Javaheripour K Muller A Degner D Wilhelm J et al Hyperhomocysteinemia as a new risk factor for brain shrinkage in patients with alcoholism. Neurosci Lett (2003) 335(3):179–82. 10.1016/s0304-3940(02)01194-1
27.
Halsted CH Villanueva JA Devlin AM Niemela O Parkkila S Garrow TA et al Folate deficiency disturbs hepatic methionine metabolism and promotes liver injury in the ethanol-fed micropig. Proc Natl Acad Sci U S A (2002) 99(15):10072–7. 10.1073/pnas.112336399
28.
Villanueva JA Halsted CH . Hepatic transmethylation reactions in micropigs with alcoholic liver disease. Hepatology (2004) 39(5):1303–10. 10.1002/hep.20168
29.
Purohit V Abdelmalek MF Barve S Benevenga NJ Halsted CH Kaplowitz N et al Role of S-adenosylmethionine, folate, and betaine in the treatment of alcoholic liver disease: Summary of a symposium. Am J Clin Nutr (2007) 86(1):14–24. 10.1093/ajcn/86.1.14
30.
Ji C Nagaoka K Zou J Casulli S Lu S Cao KY et al Chronic ethanol-mediated hepatocyte apoptosis links to decreased TET1 and 5-hydroxymethylcytosine formation. FASEB J (2019) 33(2):1824–35. 10.1096/fj.201800736R
31.
Tammen SA Park JE Shin PK Friso S Chung J Choi S-W . Iron supplementation reverses the reduction of hydroxymethylcytosine in hepatic DNA associated with chronic alcohol consumption in rats. J Cancer Prev (2016) 21(4):264–70. 10.15430/JCP.2016.21.4.264
32.
Tammen SA Park LK Dolnikowski GG Ausman LM Friso S Choi S-W . Hepatic DNA hydroxymethylation is site-specifically altered by chronic alcohol consumption and aging. Eur J Nutr (2017) 56(2):535–44. 10.1007/s00394-015-1098-4
33.
Villanueva JA Esfandiari F White ME Devaraj S French SW Halsted CH . S-adenosylmethionine attenuates oxidative liver injury in micropigs fed ethanol with a folate-deficient diet. Alcohol Clin Exp Res (2007) 31(11):1934–43. 10.1111/j.1530-0277.2007.00511.x
34.
Kharbanda KK Rogers DD II Mailliard ME Siford GL Barak AJ Beckenhauer HC et al A comparison of the effects of betaine and S-adenosylmethionine on ethanol-induced changes in methionine metabolism and steatosis in rat hepatocytes. J Nutr (2005) 135(3):519–24. 10.1093/jn/135.3.519
35.
Barbier E Tapocik JD Juergens N Pitcairn C Borich A Schank JR et al DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci (2015) 35(15):6153–64. 10.1523/JNEUROSCI.4571-14.2015
36.
Cervera-Juanes R Wilhelm LJ Park B Grant KA Ferguson B . Alcohol-dose-dependent DNA methylation and expression in the nucleus accumbens identifies coordinated regulation of synaptic genes. Transl Psychiatry (2017) 7(1):e994. 10.1038/tp.2016.266
37.
Vendruscolo LF Barbier E Schlosburg JE Misra KK Whitfield TW Jr Logrip ML et al Corticosteroid-dependent plasticity mediates compulsive alcohol drinking in rats. J Neurosci (2012) 32(22):7563–71. 10.1523/JNEUROSCI.0069-12.2012
38.
Heilig M Barbier E Johnstone AL Tapocik J Meinhardt MW Pfarr S et al Reprogramming of mPFC transcriptome and function in alcohol dependence. Genes, Brain Behav (2016) 16(1):86–100. 10.1111/gbb.12344
39.
Taqi MM Bazov I Watanabe H Sheedy D Harper C Alkass K et al Prodynorphin CpG-SNPs associated with alcohol dependence: Elevated methylation in the brain of human alcoholics. Addict Biol (2011) 16(3):499–509. 10.1111/j.1369-1600.2011.00323.x
40.
Kyzar EJ Zhang H Sakharkar AJ Pandey SC . Adolescent alcohol exposure alters lysine demethylase 1 (LSD1) expression and histone methylation in the amygdala during adulthood. Addict Biol (2017) 22(5):1191–204. 10.1111/adb.12404
41.
Palmisano M Pandey SC . Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol (2017) 60:7–18. 10.1016/j.alcohol.2017.01.001
42.
Moonat S Starkman BG Sakharkar A Pandey SC . Neuroscience of alcoholism: Molecular and cellular mechanisms. Cel Mol Life Sci (2009) 67:73–88. 10.1007/s00018-009-0135-y
43.
Kyzar EJ Bohnsack JP Zhang H Pandey SC . MicroRNA-137 drives epigenetic reprogramming in the adult amygdala and behavioral changes after adolescent alcohol exposure. eNeuro (2019) 6(6):ENEURO.0401–19.2019. 10.1523/ENEURO.0401-19.2019
44.
Bohnsack JP Teppen T Kyzar EJ Dzitoyeva S Pandey SC . The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl Psychiatry (2019) 9(1):34. 10.1038/s41398-019-0367-z
45.
Kyzar EJ Zhang H Pandey SC . Adolescent alcohol exposure epigenetically suppresses amygdala Arc enhancer RNA expression to confer adult anxiety susceptibility. Biol Psych (2019) 85(11):904–14. 10.1016/j.biopsych.2018.12.021
46.
Qiang M Denny A Chen J Ticku MK Yan B Henderson G . The site specific demethylation in the 5′-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription. PLoS ONE (2010) 5(1):e8798. 10.1371/journal.pone.0008798
47.
Bleich S Degner D Sperling W Bonsch D Thurauf N Kornhuber J . Homocysteine as a neurotoxin in chronic alcoholism. Prog Neuropsychopharmacol Biol Psychiatry (2004) 28(3):453–64. 10.1016/j.pnpbp.2003.11.019
48.
Bonsch D Lenz B Kornhuber J Bleich S . DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. NeuroReport (2005) 16(2):167–70. 10.1097/00001756-200502080-00020
49.
Rossetti ZL Melis F Carboni S Gessa GL . Marked decrease of extraneuronal dopamine after alcohol withdrawal in rats: Reversal by MK-801. Euro J Pharm (1991) 200(2-3):371–2. 10.1016/0014-2999(91)90600-u
50.
Salinas AG Mateo Y Cuzon Carlson VC Stinnett GS Luo G Seasholtz AF et al Long-term alcohol consumption alters dorsal striatal dopamine release and regulation by D2 dopamine receptors in rhesus macaques. Neuropsychopharmacology (2021) 46:1432–41. 10.1038/s41386-020-00938-8
51.
Siciliano CA Karkhanis AN Holleran KM Melchior JR Jones SR . Cross-species alterations in synaptic dopamine regulation after chronic alcohol exposure. In: GrantKLovingerD, editors. The neuropharmacology of alcohol. Cham: Springer (2018). p. 213–38. 10.1007/164_2018_106
52.
Dobbs LK Lemos JC Alvarez VA . Restructuring of basal ganglia circuitry and associated behaviors triggered by low striatal D2 receptor expression: Implications for substance use disorders. Genes, Brain Behav (2016) 16(1):56–70. 10.1111/gbb.12361
53.
Bidwell LC Karoly HC Thayer RE Claus ED Bryan AD Weiland BJ et al DRD2 promoter methylation and measures of alcohol reward: Functional activation of reward circuits and clinical severity. Addict Biol (2019) 24(3):539–48. 10.1111/adb.12614
54.
Budygin EA Oleson EB Mathews TA Lack AK Diaz MR McCool BA et al Effects of chronic alcohol exposure on dopamine uptake in rat nucleus accumbens and caudate putamen. Psychopharmacology (2007) 193:495–501. 10.1007/s00213-007-0812-1
55.
Jasiewicz A Rubis B Samochowiec J Malecka I Suchanecka A Jablonski M et al DAT1 methylation changes in alcohol-dependent individuals vs. controls. J Psychiatr Res (2015) 64:130–3. 10.1016/j.jpsychires.2015.03.007
56.
Jiao X Pare WP Tejani-Butt SM . Alcohol consumption alters dopamine transporter sites in Wistar-Kyoto rat brain. Brain Res (2006) 1073-1074:175–82. 10.1016/j.brainres.2005.12.009
57.
Grover T Gupta R Arora G Shekhar Bal C Ambekar A Basu Ray S et al Dopamine transporter availability in alcohol and opioid dependent subjects – A 99mTc-TRODAT-1SPECT imaging and genetic association study. Psychiatry Res Neuroimaging (2020) 305:111187. 10.1016/j.pscychresns.2020.111187
58.
Gupta R Singh R Ambekar A Grover T Sharma A . M112 – a study on the association of polymorphisms and methylation status of dopamine pathway genes in alcohol dependence. Euro Neuropsychopharmacol (2019) 29:S1016. 10.1016/j.euroneuro.2017.08.419
59.
Hillemacher T Frieling H Hartl T Wilhelm J Kornhuber J Bleich S . Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J Psychiatr Res (2009) 43(4):388–92. 10.1016/j.jpsychires.2008.04.006
60.
Wiers CE Shumay E Volkow ND Frieling H Kotsiari A Lindenmeyer J et al Effects of depressive symptoms and peripheral DAT methylation on neural reactivity to alcohol cues in alcoholism. Transl Psychiatry (2015) 5(9):e648. 10.1038/tp.2015.141
61.
Mittleman G Call SB Cockroft JL Goldowitz D Matthews DB Blaha CD . Dopamine dynamics associated with, and resulting from, schedule-induced alcohol self-administration: Analyses in dopamine transporter knockout mice. Alcohol (2011) 45(4):325–39. 10.1016/j.alcohol.2010.12.006
62.
Laine TP Ahonen A Torniainen P Heikkila J Pyhtinen J Rasanen P et al Dopamine transporters increase in human brain after alcohol withdrawal. Mol Psychiatry (1999) 4:189–91. 10.1038/sj.mp.4000514
63.
Yoshimoto K Ueda S Nishi M Yang Y Matsushita H Takeuchi Y et al Changes in dopamine transporter and c‐Fos expression in the nucleus accumbens of alcohol‐tolerant rats. Alc Clin Exp Res (2006) 24(3):361–5. 10.1111/j.1530-0277.2000.tb04623.x
64.
Gatta E Grayson DR Auta J Saudagar V Dong E Chen Y et al Genome-wide methylation in alcohol use disorder subjects: Implications for an epigenetic regulation of the cortico-limbic glucocorticoid receptors (NR3C1). Mol Psychiatry (2021) 26(3):1029–41. 10.1038/s41380-019-0449-6
65.
Richardson HN Lee SY O'Dell LE Koob GF River CL . Alcohol self-administration acutely stimulates the hypothalamic-pituitary-adrenal axis, but alcohol dependence leads to a dampened neuroendocrine state. Eur J Neurosci (2008) 28(8):1641–53. 10.1111/j.1460-9568.2008.06455.x
66.
Stephens MAC Wand G . Stress and the HPA axis: Role of glucocorticoids in alcohol dependence. Alcohol Res (2012) 34(4):468–83.
67.
Kozuka T Omori Y Watanabe S Tarusawa E Yamamoto H Chaya T et al miR-124 dosage regulates prefrontal cortex function by dopaminergic modulation. Sci Rep (2019) 9(1):3445. 10.1038/s41598-019-38910-2
68.
Hashimoto H Vertino PM Cheng X . Molecular coupling of DNA methylation and histone methylation. Epigenomics (2010) 2(5):657–69. 10.2217/epi.10.44
69.
Rose NR Klose RJ . Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta (2014) 1839(12):1362–72. 10.1016/j.bbagrm.2014.02.007
70.
Du J Johnson LM Jacobsen SE Patel DJ . DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cel Biol (2015) 16:519–32. 10.1038/nrm4043
71.
Bannister AJ Kouzarides T . Reversing histone methylation. Nature (2005) 436(7054):1103–6. 10.1038/nature04048
72.
Klose RJ Zhang Y . Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cel Biol (2007) 8:307–18. 10.1038/nrm2143
73.
Lan F Nottke AC Shi Y . Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cel Biol (2008) 20(3):316–25. 10.1016/j.ceb.2008.03.004
74.
Black JC Van Rechem C Whetstine JR . Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol Cel (2012) 48(4):491–507. 10.1016/j.molcel.2012.11.006
75.
Hyun K Jeon J Park K Kim J . Writing, erasing and reading histone lysine methylations. Exp Mol Med (2017) 49(4):e324. 10.1038/emm.2017.11
76.
Shinkai Y Tachibana M . H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev (2011) 25(8):781–8. 10.1101/gad.2027411
77.
Klose RJ Kallin EM Zhang Y . JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet (2006) 7(9):715–27. 10.1038/nrg1945
78.
Kouzarides T . Histone methylation in transcriptional control. Curr Opin Genet Develop (2002) 12(2):198–209. 10.1016/s0959-437x(02)00287-3
79.
Pal-Bhadra M Bhadra U Jackson DE Mamatha L Park P-H Shukla SD . Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sci (2007) 81(12):979–87. 10.1016/j.lfs.2007.07.030
80.
Shukla SD Lim RW . Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res (2013) 35(1):47–55.
81.
Page A Paoli PP Hill SJ Howarth R Wu R Kweon S-M et al Alcohol directly stimulates epigenetic modifications in hepatic stellate cells. J Hepatol (2015) 62(2):388–97. 10.1016/j.jhep.2014.09.033
82.
D'Addario C Caputi FF Ekstrom TJ Benedetto MD Maccarrone M Romualdi P et al Ethanol induces epigenetic modulation of prodynorphin and pronociceptin gene expression in the rat amygdala complex. J Mol Neurosci (2013) 49(2):312–9. 10.1007/s12031-012-9829-y
83.
Blaine SK Sinha R . Alcohol, stress, and glucocorticoids: From risk to dependence and relapse in alcohol use disorders. Neuropharmacology (2017) 122:136–47. 10.1016/j.neuropharm.2017.01.037
84.
Frias J Torres JM Miranda MT Ruiz E Ortega E . Effects of acute alcohol intoxication on pituitary-gonadal axis hormones, pituitary-adrenal axis hormones, β-endorphin and prolactin in human adults of both sexes. Alcohol Alcohol (2002) 37(2):169–73. 10.1093/alcalc/37.2.169
85.
Bohnsack JP Zhang H Wandling GM He D Kyzar EJ Lasek AW et al Targeted epigenomic editing ameliorates adult anxiety and excessive drinking after adolescent alcohol exposure. Sci Adv (2022) 8(18):abn2748. 10.1126/sciadv.abn2748
86.
Berkel TD Zhang H Teppen T Sakharkar AJ Pandey SC . Essential role of histone methyltransferase G9a in rapid tolerance to the anxiolytic effects of ethanol. Int J Neuropsychopharmacol (2018) 22(4):292–302. 10.1093/ijnp/pyy102
87.
Qiang M Denny A Lieu M Carreon S Li J . Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics (2011) 6(9):1095–104. 10.4161/epi.6.9.16924
88.
Zeng K Xie A Zhang X Zhong B Liu X Hao W . Chronic alcohol treatment-induced GABA-Aα5 histone H3K4 trimethylation upregulation leads to increased GABA-Aα5 expression and susceptibility to alcohol addiction in the offspring of Wistar rats. Front Psychiatry (2018) 9:468. 10.3389/fpsyt.2018.00468
89.
Enoch MA . The role of GABAA receptors in the development of alcoholism. Pharmacol Biochem Behav (2008) 90(1):95–104. 10.1016/j.pbb.2008.03.007
90.
Chester JA Cunningham CL . GABAA receptor modulation of the rewarding and aversive effects of ethanol. AlcohoI (2002) 26(3):131–43. 10.1016/s0741-8329(02)00199-4
91.
Wang H Ji B Ren H Meng C . The relationship between lysine 4 on histone H3 methylation levels of alcohol tolerance genes and changes of ethanol tolerance in Saccharomyces cerevisiae. Microb Biotechnol (2014) 7(4):307–14. 10.1111/1751-7915.12121
92.
Pinzón JH Reed AR Shalaby NA Buszczak M Rodan AR Rothenfluh A . Alcohol-induced behaviors require a subset of drosophila JmjC-domain histone demethylases in the nervous system. Alcohol Clin Exp Res (2017) 41(12):2015–24. 10.1111/acer.13508
93.
Ramirez-Roman ME Billini CE Ghezzi A . Epigenetic mechanisms of alcohol neuroadaptation: Insights from Drosophila. J Exp Neurosci (2018) 12:1179069518779809. 10.1177/1179069518779809
94.
Malovic E Ealy A Kanthasamy A Kanthasamy AG . Emerging roles of N6-methyladenosine (m6A) epitranscriptomics in toxicology. Toxicol Sci (2021) 181(1):13–22. 10.1093/toxsci/kfab021
95.
Malovic E Pandey SC . N6-methyladenosine (m6A) epitranscriptomics in synaptic plasticity and behaviors. Neuropsychopharmacology (2022) 48:221–2. 10.1038/s41386-022-01414-1
96.
Zhao BS Roundtree IA He C . Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cel Biol (2017) 18(1):31–42. 10.1038/nrm.2016.132
97.
Fu Y Dominissini D Rechavi G He C . Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet (2014) 15:293–306. 10.1038/nrg3724
98.
Cao G Li H-B Yin Z Flavell RA . Recent advances in dynamic m6A RNA modification. Open Biol (2016) 6(4):160003. 10.1098/rsob.160003
99.
Meyer KD Saletore Y Zumbo P Elemento O Mason CE Jaffrey SR . Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell (2012) 149(7):1635–46. 10.1016/j.cell.2012.05.003
100.
Shi H Wang X Lu Z Zhao BS Ma H Hsu PJ et al YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res (2017) 27(3):315–28. 10.1038/cr.2017.15
101.
Zaccara S Jaffrey SR . A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell (2020) 181(7):1582–95. 10.1016/j.cell.2020.05.012
102.
Lasman L Krupalnik V Viukov S Mor N Aguilera-Castrejon A Schneir D et al Context-dependent functional compensation between Ythdf m6A reader proteins. Genes Dev (2020) 34(19-20):1373–91. 10.1101/gad.340695.120
103.
Li Y Bedi RK Moroz-Omori EV Caflisch A . Structural and dynamic insights into redundant function of YTHDF proteins. J Chem Inf Model (2020) 60(12):5932–5. 10.1021/acs.jcim.0c01029
104.
Xu K Sun Y Sheng B Zheng Y Wu X Xu K . Role of identified RNA N6-methyladenosine methylation in liver. Anal Biochem (2019) 578:45–50. 10.1016/j.ab.2019.05.005
105.
Zhao Z Meng J Su R Zhang J Chen J Ma X et al Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J Hepatol (2020) 73(3):664–79. 10.1016/j.jhep.2020.04.009
106.
Hu Y Feng Y Zhang L Jia Y Cai D Qian S-B et al GR-mediated FTO transactivation induces lipid accumulation in hepatocytes via demethylation of m 6 A on lipogenic mRNAs. RNA Biol (2020) 17(7):930–42. 10.1080/15476286.2020.1736868
107.
Yu J-T Hu X-W Chen H-Y Yang Q Li H-D Dong Y-H et al DNA methylation of FTO promotes renal inflammation by enhancing m6A of PPAR-α in alcohol-induced kidney injury. Pharmacol Res (2021) 163:105286. 10.1016/j.phrs.2020.105286
108.
Wang L Liu X Luo X Zeng M Zuo L Wang K-S . Genetic variants in the fat mass- and obesity-associated (FTO) gene are associated with alcohol dependence. J Mol Neurosci (2013) 51(2):416–24. 10.1007/s12031-013-0044-2
109.
Goodyear K Lee MR Schwandt ML Hodgkinson CA Leggio L . Hepatic, lipid and genetic factors associated with obesity: Crosstalk with alcohol dependence?World J Biol Psychiatry (2017) 18(2):120–8. 10.1080/15622975.2016.1249952
110.
Corella D Ortega-Azorin C Sorli JV Covas MI Carrasco P Salas-Salvado J et al Statistical and biological gene-lifestyle interactions of MC4R and FTO with diet and physical activity on obesity: New effects on alcohol consumption. PLoS ONE (2012) 7(12):e52344. 10.1371/journal.pone.0052344
111.
Sobczyk-Kopciol A Broda G Wojnar M Kurjata P Jakubczyk A Klimkiewicz A et al Inverse association of the obesity predisposing FTO rs9939609 genotype with alcohol consumption and risk for alcohol dependence. Addiction (2011) 106(4):739–48. 10.1111/j.1360-0443.2010.03248.x
112.
Hubacek JA Adamkova V Dlouha D Jirsa M Sperl J Tonjes A et al Fat mass and obesity-associated (fto) gene and alcohol intake. Addiction (2012) 107(6):1185–6. 10.1111/j.1360-0443.2012.03825.x
113.
Rask-Andersen M Karlsson T Ek WE Johansson A . Gene-environment interaction study for BMI reveals interactions between genetic factors and physical activity, alcohol consumption and socioeconomic status. Plos Genet (2017) 13(9):e1006977. 10.1371/journal.pgen.1006977
114.
Lichenstein SD Jones BL O'Brien JW Zezza N Stiffler S Holmes B et al Familial risk for alcohol dependence and developmental changes in BMI: The moderating influence of addiction and obesity genes. Pharmacogenomics (2014) 15(10):1311–21. 10.2217/pgs.14.86
115.
Hess ME Hess S Meyer KD Verhagen LA Koch L Bronneke HS et al The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci (2013) 16:1042–8. 10.1038/nn.3449
116.
Chen X Yu C Guo M Zheng X Ali S Huang H et al Down-regulation of m6A mRNA methylation Is involved in dopaminergic neuronal death. ACS Chem Neurosci (2019) 10(5):2355–63. 10.1021/acschemneuro.8b00657
117.
Sevgi M Rigoux L Kuhn AB Mauer J Schilbach L Hess ME et al An obesity-predisposing variant of the FTO gene regulates D2R-dependent reward learning. J Neurosci (2015) 35(36):12584–92. 10.1523/JNEUROSCI.1589-15.2015
118.
Xue A Huang Y Li M Wei Q Bu Q . Comprehensive analysis of differential m6A RNA methylomes in the hippocampus of cocaine-conditioned mice. Mol Neurobiol (2021) 58:3759–68. 10.1007/s12035-021-02363-4
119.
Wiemerslage L Nilsson EK Dahlberg LS Ence-Eriksson F Castillo S Larsen AL et al An obesity‐associated risk allele within the FTO gene affects human brain activity for areas important for emotion, impulse control and reward in response to food images. Eur J Neurosci (2016) 43(9):1173–80. 10.1111/ejn.13177
120.
Clarke T-K Treutlein J Zimmermann US Kiefer F Skowronek MH Rietschel M et al HPA-axis activity in alcoholism: Examples for a gene-environment interaction. Addict Biol (2008) 13(1):1–14. 10.1111/j.1369-1600.2007.00084.x
121.
Engel M Eggert C Kaplick PM Eder M Roh S Tietze L et al The role of m6A/m-RNA methylation in stress response regulation. Neuron (2018) 99(2):389–403. 10.1016/j.neuron.2018.07.009
Summary
Keywords
alcohol use disorder, brain, epigenetics, DNA methylation, histone methylation, RNA methylation
Citation
Cruise TM, Kotlo K, Malovic E and Pandey SC (2023) Advances in DNA, histone, and RNA methylation mechanisms in the pathophysiology of alcohol use disorder. Adv. Drug. Alco. Res. 3:10871. doi: 10.3389/adar.2023.10871
Received
29 August 2022
Accepted
25 January 2023
Published
15 February 2023
Volume
3 - 2023
Edited by
Emmanuel Onaivi, William Paterson University, United States
Reviewed by
Dipak Sarkar, Rutgers, The State University of New Jersey, United States
Youssef Sari, University of Toledo, United States
Updates
Copyright
© 2023 Cruise, Kotlo, Malovic and Pandey.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subhash C. Pandey, scpandey@uic.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.