Abstract
Background: There are isolated reports of mutations in genes for isocitrate dehydrogenases (IDH1 and IDH2), but few have been examined in a large number of different malignancies. We aimed to analyze mutational prevalence of these genes in a large series of cancers and determine their significance in most mutated phenotype.
Methods: We analyzed the frequencies of IDH1 and IDH2 mutations in 14,726 malignancies of 37 cancers. Furthermore, we examined these mutations in the most frequent cancer (gliomas, 923 cases) from a single cohort, and determined their clinical significance.
Results: IDH1 mutations were present in 3% (473/14,726) of cancers. The highest frequencies were in oligodendrogliomas (91/102, 89%), anaplastic oligodendrogliomas (40/46, 87%), and diffuse astrocytomas (89/116, 77%). IDH2 mutation was detected in <1% (83/14,726) of cancers, but were present in 13% (6/46) of anaplastic oligodendrogliomas, 9% (9/102) of oligodendrogliomas, and in 5% (2/39) of cutaneous squamous cell carcinomas. Further analyses of 923 gliomas revealed 34 and 1% of IDH1 and IDH2 mutations, respectively. In up to 342 months of follow-up, IDH1 and IDH2 mutations were significantly linked with better overall (OS) (both p = 0.01) and progression-free survival (PFS) (p = 0.01; p = 0.004), respectively.
Conclusion: IDH1 and IDH2 are often mutated in a tissue-specific manner, most commonly in gliomas. Mutation in both genes is linked to OS and PFS. Our findings suggest that these genes are promising therapeutic targets and strong prognostic biomarkers in gliomas.
Introduction
Malignancy is the most common non-communicable disease and accountable for one in eight deaths across the globe. The incidence of cancer and cancer-related death for the year 2021 has been projected to occur in 1,898,160 and 608,570 cases, respectively in the United States. The cancer-related death rate has significantly fallen since 2017, accounting for an overall decline of 29% that is equal to 2.9 million fewer deaths. Over the past decade, the death rate was shown to be significantly decreased in the leading cancers including lung, colorectal, breast, and prostate nevertheless, the decline slowed in breast and colorectal cancers of females, and the decline stopped in prostate cancer (1). Various next-generation transcriptomic, genomic, and proteomic studies identified many genetically deregulated genes in human cancer. These altered cancer gene clusters exert deregulated signaling on certain pathways as somatic mutations in receptor tyrosine kinases (RTKs) and their downstream pathway members including different RAS (H/K/N) molecules constitutively trigger canonical mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) signaling. Particularly, high frequent genetic alterations of various genes like TP53, BRAF, RAS, EGFR, PIK3CA, PTEN, HER2, UDX, ALK, TERT, mTOR; IDH1, etc. were documented in the oncogenesis of several kinds of human malignancies (2–17). Aberrant activation of the vital pathways promotes uncontrolled cell division, proliferation, growth, invasion, and metastasis that collectively leads to tumorigenesis.
Isocitrate dehydrogenase 1 (IDH1) gene mutations were high frequently detected in human cancers particularly in secondary glioblastomas (>70%) (18). IDH1 mutations are present mainly in the hotspot arginine at codon R132 and many different IDH1 mutations (R132H, R132S, R132C, and R132G) were also reported for the residue R132. The IDH2 mutations were identified in codon 172 and malignancies with no mutations in IDH1 frequently showed mutations in the cognate amino acid arginine (R) at 172 of the IDH2 gene. All the codon R132 IDH1 mutants were shown to decrease the enzymatic activity of the IDH1 (16). Frequent mutations of the IDH1 have also been identified in hormone receptor-positive (HR+) breast adenocarcinoma, thyroid cancer, cholangiocarcinoma (50–70%), and IDH2 (R172S) in benign giant cell tumors of the bone (80%) (17, 19–22).
IDH plays a key role within the Krebs cycle and produces alpha-ketoglutarate (α-KG) by catalyzing the oxidative decarboxylation of isocitrate. The IDH activity is exclusively dependent on nicotinamide adenine dinucleotide phosphate (NADP+) which is catalyzed by IDH1 to produce NADPH that is involved in controlling oxidative damage of a cell (23). The cancer-associated IDH1 mutations have been demonstrated to produce 2-hydroxyglutarate as this IDH (IDH1 and IDH2) mutant enzyme carries a neomorphic catalytic function and converts alpha-ketoglutarate to 2-hydroxyglutarate that suppresses the histone lysine demethylases (23–25). It has been reported that an IDH1 mutation was potentially able to form glioma hypermethylation phenotype while IDH2 could promote acute myeloid leukemia (26). Nevertheless, a prominent status of the IDH1 and IDH2 mutations has never been undertaken particularly within a large number of different human cancer samples.
Given the importance of these genes, we investigated the IDH1 and IDH2 mutations in a large series of cancer cases (n = 14,726) (solid malignancies) from the data of The Cancer Genome Atlas and the Memorial Slone Kettering Cancer Centre.
Methods
As detailed in Table 1, we analyzed 15,300 malignant tumour samples (obtained from 14,726 patients) of 37 different types of malignancies in solid tissue cancers. All data was derived from The Cancer Genome Atlas studies by performing various analyses using the methods within the cBioPortal (www.cbioportal.org), an open-access, open-source, and publicly available platform for interactive exploration of multidimensional cancer genomics datasets. Approval statement/informed consent is not required for this study as we used data from a publicly available database.
TABLE 1
| S. No | Name of organ | Type of cancer | Number of samples |
|---|---|---|---|
| 1 | Adrenal gland | Adrenocortical carcinoma | 92 |
| 2 | Ampulla of vater | Ampullary carcinoma | 160 |
| 3 | Biliary tract | Colangiocarcinoma | 195 |
| 4 | Bladder | Bladder cancer | 413 |
| 5 | Bowel | Colorectal adenocarcinoma | 619 |
| 6 | Breast | Breast cancer | 2,509 |
| 7 | Brain | Glioma | 1,004 |
| 8 | Glioblastoma | 543 | |
| 9 | Medulloblastoma | 125 | |
| 10 | Cervix | Cervical squamous cell carcinoma | 297 |
| 11 | Esophagus | Esophageal squamous cell carcinoma | 139 |
| 12 | Esophageal carcinoma | 559 | |
| 13 | Gastric adenocarcinoma | 78 | |
| 14 | Metastastic esophagogastric cancer | 341 | |
| 15 | Esophageal adenocarcinoma | 182 | |
| 16 | Stomach | Stomach adenocarcinoma | 440 |
| 17 | Eye | Uveal melanoma | 80 |
| 18 | Head and Neck | Head and neck squamous cell carcinoma | 523 |
| 19 | Oral squamous cell carcinoma | 40 | |
| 20 | Nasopharygeal carcinoma | 56 | |
| 21 | Kidney | Kidney renal clear cell carcinoma | 446 |
| 22 | Liver | Hepatocellular Adenoma | 46 |
| 23 | Hepatocellular carcinomas | 243 | |
| 24 | Lung | Small cell lung cancer | 120 |
| 25 | Non-small cell lung cancer | 447 | |
| 26 | Ovary | Ovarian serous cystadenocarcinoma | 489 |
| 27 | Pancreas | Pancreatic adenocarcinoma | 456 |
| 28 | Peripheral nervous system | Pediatric neuroblastoma | 1,089 |
| 29 | Pleura | Pleural mesothelioma | 22 |
| 30 | Prostate | Metastatic prostate adenocarcinoma | 444 |
| 31 | Prostate adenocarcinoma | 1,465 | |
| 32 | Skin | Basel cell carcinoma | 293 |
| 33 | Cutaneous squamous cell carcinoma | 39 | |
| 34 | Metastatic melanoma | 110 | |
| 35 | Skin cutaneous melanoma | 448 | |
| 36 | Testis | Germ cell tumors | 180 |
| 37 | Thymus | Thymoma | 123 |
| 38 | Uterus | Uterine corpus endometrial carcinoma | 373 |
| 39 | Uterine carcinosarcoma | 57 | |
| 40 | Vulva | Squamous cell carcinoma of the vulva | 15 |
| Total | 15,300a |
Human cancer samples (solid tumors) analyzed for IDH1 and IDH2 gene mutations.
Derived from 14,726 patients.
We explored the cBioPortal database for mutational analysis of IDH1 and IDH2. In brief, firstly we selected one/two cohorts of larger sample size against each malignancy from the different cohorts (cancer studies) available in the datasets (accessible from the homepage of cBioPortal). In total, 40 different cohorts were selected (users have the option to select one or more than one cohort). Secondly, we selected only “mutation” under the genomic profile menu (on the same page). Other genomic profiles including the copy number variants (CNVs), structural variant, RNAseq, etc., were excluded (unselected). Thirdly, we selected “samples with mutation data” under the patient/case set menu. Fourthly, we input IDH1 or IDH2 under the “enter genes menu” (users have options to enter multiple genes) and queried the IDH1/IDH2 against the selected set of malignancies. The query generated a newer window with multiple tool tab options such as oncoprint, cancer types summary, plots, mutations, survival, etc., to visualize the results against various parameters of selected cohorts. Users may perform different analyses of selected datasets by selecting and submitting the intended (any of the above-indicated) tool tab. We utilized the tool tab “cancer types summary” to visualize and obtain the prevalence of IDH1/IDH2 mutations in various selected malignancies. Complete and comprehensive step-by-step procedures are clearly described previously for mining the cBioPortal database (27).
The Memorial Sloan Kettering Cancer Centre data of 1,004 glioma samples (derived from 923 patients) was analyzed for IDH1 and IDH2 mutations within the cBioPortal (www.cbioportal.org), and clinical links including overall survival (OS) and progression-free survival (PFS) was performed by selecting the tool tab “survival tab,” excluded the overlapping samples and results were visualized as described earlier (27).
Statistical genomic analyses were executed utilizing the methods incorporated within the cBioPortal (www.cbioportal.org) (27). Kaplan–Meier plots with a log-rank test were performed to determine the OS and PFS of gliomas presence of a minimum of one mutation or absence of mutation in the related queried candidate gene. p < 0.05 was regarded as statistically significant.
Results
The composition of the 15,300 different cancer samples and their malignant types are shown in Table 1. As detailed in Table 2, overall, 3% (473/14,726) of solid tumour cancer cases harbored mutations in IDH1. The highest frequencies were present in oligodendrogliomas, anaplastic oligodendrogliomas, and diffuse astrocytomas. The overall prevalence of IDH2 mutation was <1% (83/14,726). The highest frequencies were present in anaplastic oligodendrogliomas, oligodendrogliomas, and cutaneous squamous cell carcinomas. These results clearly indicate that IDH1 and IDH2 are often mutated in solid cancers in a tissue-specific manner and the mutational incidence was frequent mainly in gliomas, suggesting that IDH1 and IDH2 may play important roles in these types of solid human cancer.
TABLE 2
| Type of cancer | Mutational prevalence of IDH1a | Type of cancer | Mutational prevalence of IDH2a |
|---|---|---|---|
| Oligodendroglioma | 89.2% (91/102) | Anaplastic Oligodendroglioma | 13.0% (6/46) |
| Anaplastic Oligodendroglioma | 87.0% (40/46) | Oligodendroglioma | 8.8% (9/102) |
| Diffuse Astrocytoma | 76.7% (89/116) | Cutaneous Squamous Cell Carcinoma | 5.1% (2/39) |
| Anaplastic Astrocytoma | 54.1% (86/159) | Skin Cancer, Non-Melanoma | 3.4% (10/293) |
| Intrahepatic Cholangiocarcinoma | 29.1% (46/158) | Lung Squamous Cell Carcinoma | 2.6% (4/155) |
| Glioblastoma Multiforme | 5.6% (45/796) | Intrahepatic Cholangiocarcinoma | 2.5% (4/158) |
| Extrahepatic Cholangiocarcinoma | 5.4% (2/37) | Uterine Endometrioid Carcinoma | 2.0% (4/200) |
| Cutaneous Squamous Cell Carcinoma | 5.1% (2/39) | Colorectal Adenocarcinoma | 1.8% (11/619) |
| Cutaneous Melanoma | 4.9% (27/550) | Uterine Carcinosarcoma | 1.7% (1/57) |
| Skin Cancer, Non-Melanoma | 3.7% (11/293) | Diffuse Astrocytoma | 1.7% (2/116) |
| Esophagogastric Adenocarcinoma | 2.5% (2/80) | Tubular Stomach Adenocarcinoma | 1.3% (1/79) |
| Uterine Serous Carcinoma | 2.3% (1/44) | Anaplastic Astrocytoma | 1.3% (2/159) |
| Bladder Urothelial Carcinoma | 2.2% (9/412) | Esophagogastric Adenocarcinoma | 1.2% (1/80) |
| Colorectal Adenocarcinoma | 2.1% (13/619) | Hepatocellular Carcinoma | 1.2% (3/243) |
| Adenocarcinoma of the Gastroesophageal Junction | 1.7% (1/57) | Esophagogastric Cancer | 1.1% (4/347) |
| Uterine Endometrioid Carcinoma | 1.5% (3/200) | Esophageal Squamous Cell Carcinoma | 0.9% (3/324) |
| Stomach Adenocarcinoma | 1.3% (4/308) | Esophageal Adenocarcinoma | 0.76% (2/303) |
| Tubular Stomach Adenocarcinoma | 1.3% (1/79) | Glioblastoma Multiforme | 0.5% (4/796) |
| Esophagogastric Cancer | 1.1% (4/347) | Bladder Urothelial Carcinoma | 0.5% (2/412) |
| Thymoma | 0.8% (1/123) | Cervical Squamous Cell Carcinoma | 0.4% (1/247) |
| Head and Neck Squamous Cell Carcinoma | 0.8% (4/515) | Prostate Adenocarcinoma | 0.4% (7/1909) |
| Ampullary Carcinoma | 0.6% (1/160) | Cutaneous Melanoma | 0.4% (2/550) |
| Esophageal Squamous Cell Carcinoma | 0.6% (2/324) | Stomach Adenocarcinoma | 0.3% (1/308) |
| Renal Clear Cell Carcinoma | 0.5% (2/426) | Head and Neck Squamous Cell Carcinoma | 0.2% (1/515) |
| Prostate Adenocarcinoma | 0.4% (8/1909) |
Mutational prevalence of IDH1 and IDH2 genes in various human cancers.
% (Mutated samples/Analyzed total samples) of the indicated cancer type.
Our analysis of cancer data from The Cancer Genome Atlas revealed a high incidence of IDH1 and IDH2 mutations in subtypes of gliomas. Thus, to determine the importance of these gene mutations in glioma, we analyzed them in a single and a large cohort of glioma from the Memorial Sloan Kettering Cancer Centre data sets and also examined the association of mutations of these genes in the prognosis of gliomas. As seen in Figure 1A, IDH1 harbored somatic mutations overall in 34% (314/923) of gliomas, whilst its presence significantly predicts an improved overall survival (OS) and better progression-free survival (PFS) (Figures 1B,C). The IDH2 mutation was detected overall in 2.5% (23/923) of gliomas (Figure 2A). The IDH2 mutation-bearing patients were also statistically significantly associated with improved OS and better PFS (Figures 2B,C).
FIGURE 1
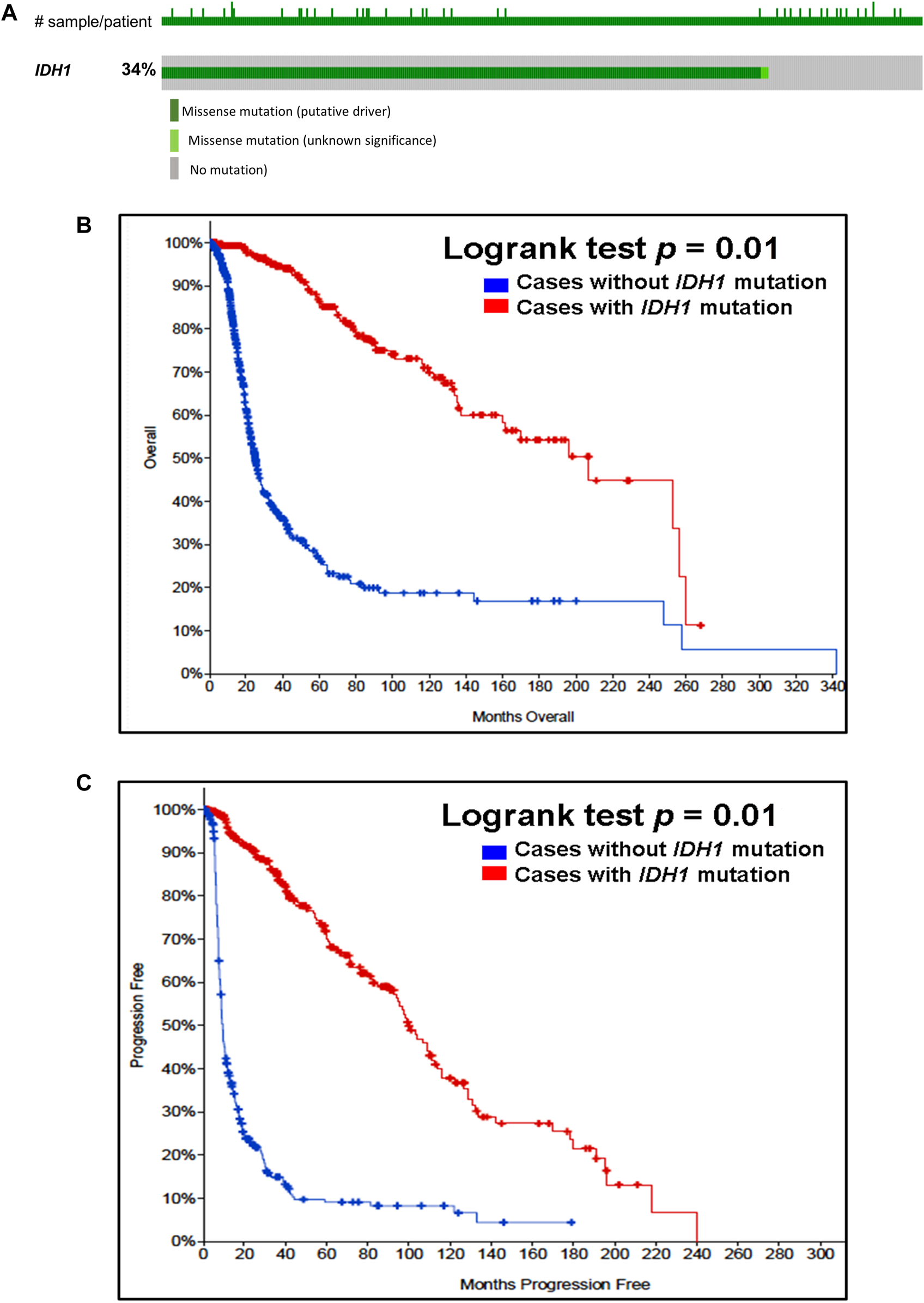
Prevalence and prognostic significance of IDH1 mutations in gliomas (A)OncoPrint tab. The tab shows the IDH1 mutations identified in gliomas. The row indicates the IDH1 gene and each column show a tumor sample. The green squares plotted on the columns show non-synonymous somatic mutations. (B)Overall survival curve. Of 923 glioma cases, 2 patients were excluded from survival analysis due to overlap. The total number of patients included in the overall survival analysis = 921. Number of cases with IDH1 mutation = 312 (number of events = 64; median overall survival (months) = 207). Number of cases without IDH1 mutation = 609 (number of events = 280; median overall survival (months) = 25), p = 0.01. (C)Progression-free survival curve. The total number of patients included in the overall progression-free survival analysis = 622. Number of cases with IDH1 mutation = 302 (number of events = 116; median progression-free survival (months) = 100). Number of cases without IDH1 mutation = 320 (number of events = 262; median progression-free survival (months) = 9), p = 0.01. Diagrams (B,C) display the Kaplan–Meier plot of overall survival and progression-free survival of glioma patients in the absence or presence of the IDH1 mutations which is indicated in blue and red colour, respectively.
FIGURE 2
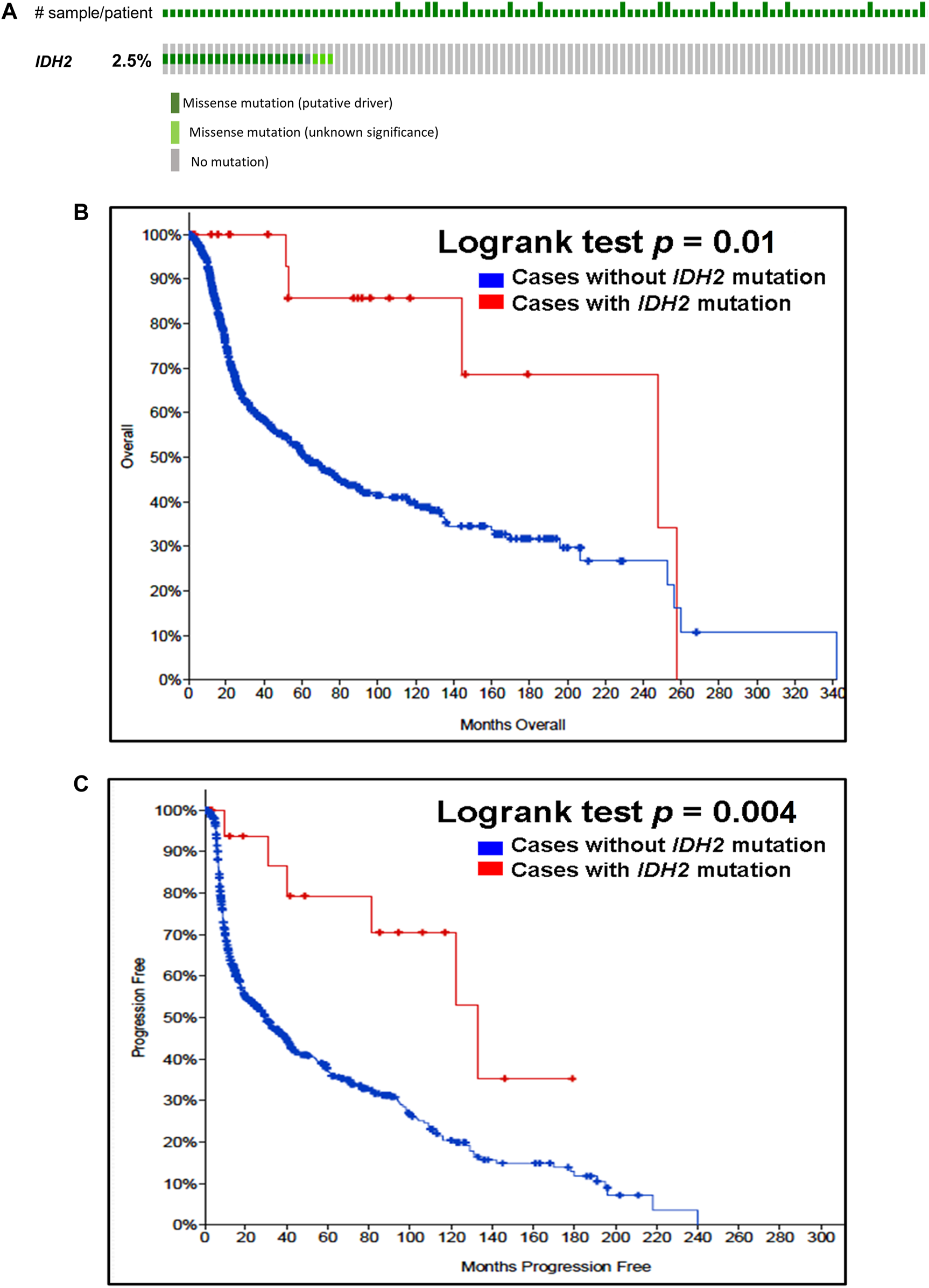
Prevalence and prognostic significance of IDH2 mutations in gliomas (A)OncoPrint tab. The tab shows the IDH2 mutations found in gliomas. The row indicates the IDH2 gene and each column shows a tumor sample. The green squares plotted on the columns show non-synonymous somatic mutations. (B)Overall survival curve. The total number of patients included in the overall survival analysis = 923. Number of cases with IDH2 mutation = 23 (number of events = 5; median overall survival (months) = 248). Number of cases without IDH2 mutation = 900 (number of events = 339; median overall survival (months) = 62), p = 0.01. (C)Progression-free survival curve. The total number of patients included in the overall progression-free survival analysis = 623. Number of cases with IDH2 mutation = 20 (number of events = 6; median progression-free survival (months) = 133). Number of cases without IDH1 mutation = 603 (number of events = 372; median progression-free survival (months) = 30), p = 0.004. Diagrams (B,C) display the Kaplan–Meier plot of overall survival and progression-free survival of glioma patients in the absence or presence of the IDH2 mutations which are indicated in blue and red colour, respectively.
Discussion
IDH1 and IDH2 are recurrently mutated in several types of human cancer (15–17, 19–22). Particularly, point mutations of these genes are major therapeutic targets and important prognostic markers in brain tumors (15, 16). Nonetheless, the prevalence of these mutations has not been analyzed in a large number of different types of human cancer despite the availability of next-generation sequencing data. Therefore, we mined the human cancer data derived from 37 different types of cancers, finding a high rate of IDH1 and IDH2 mutations in a tissue-specific manner mainly in gliomas suggesting a role for these genes in carcinogenesis. Our analysis revealed that the overall prevalence of IDH1 mutations in cancer was 3%. The oligodendrogliomas showed a high rate of IDH1 mutation, followed by anaplastic oligodendrogliomas and diffuse astrocytomas (all > 75%). In contrast, the overall prevalence of IDH2 mutation was 1%, being most frequent in anaplastic oligodendrogliomas, oligodendrogliomas and cutaneous squamous cell carcinomas (<15%). Even though a number of studies were combined and cases were different in each study in our analysis, IDH1 or IDH2 mutational frequencies of our study reflect previously published results (16).
Having found a high frequency of IDH1 and IDH2 mutations in subtypes of gliomas, we hypothesised role in long-term follow-up studies, finding that both IDH1 and IDH2 mutations are strong prognostic factors, supporting other data (15, 16). Importantly, it has been shown that IDH mutations are an independent prognostic marker of favorable outcomes (28). Thus, IDH mutations have been shown to be associated with longer survival, unlike a mutated TP53 cases (29). The IDH1 mutation has been shown to be a stand-alone favorable prognostic element in low-grade oligodendrogliomas (LOs), anaplastic oligodendrogliomas (AOs) particularly when the TP53 is not overexpressed (30). Moreover, a study investigated the prognostic value of IDH mutations in 99 secondary high-grade gliomas revealed that an IDH mutation did not associate with increased PFS although secondary anaplastic glioma patients with IDH mutation showed a significantly improved outcome (31). A previous study performed a meta-analysis of IDH1 and IDH2 mutations from 55 different studies with 9,487 glioma tumors found both mutations were independently and statistically significantly associated with better OS and PFS of glioma patients (32). Analyses of 24 different studies displayed that glioma patients with IDH mutations were associated with improved OS and PFS (33). These results provide additional evidence that collectively indicates the important roles of these genes and suggests that IDH mutations are strong prognostic markers for survival in gliomas. Conversely, recently it has been shown that the median overall survival from the first progression was not significantly different between the IDH1 mutant and wild-type group when primary and secondary glioblastomas were combined. On the other hand, the median overall survival from the initial diagnosis was significantly different (34). These findings clearly indicate that IDH1 and IDH2 could serve as potential independent diagnostic and prognostic biomarkers in these malignancies.
IDH1 and IDH2 are promising molecular targets for precision therapy not only in gliomas but also in other malignancies. Consistent with this notion, the mutant IDH1 (ivosidenib) and IDH2 (enasidenib) protein inhibitors have been initially approved by the U.S. food and drug administration (FDA) for relapsed/refractory acute myeloid leukemia (AML)-bearing IDH1 and IDH2 mutations (35). Subsequently, IDH1 inhibitor was also approved for newly diagnosed cases of AML, and currently the drug is being clinically evaluated for other cancers including cholangiocarcinoma with IDH1 mutation (35, 36). Furthermore, the IDH1-mutated tumors were recently targeted by a vaccine that exhibited vaccine-mediated tumor response in the majority of cases (37). Particularly, the FDA has already approved the mutant IDH1 and IDH2 test and hence, these findings can be expanded by testing IDH1 and IDH2 mutations in different malignancies in both the diagnostic phase and during the course of treatment to examine if the mutation evolves so that the tumors harboring IDH1 and IDH2 mutation could benefit from the IDH1/2-mediated targeted therapy. These advances collectively demonstrate that the IDH1 and IDH2 mutations play a key role in the therapeutic determination of gliomas and a subset of other malignancies.
In conclusion, we identified a high incidence of IDH1 and IDH2 mutations in a tissue-specific manner most notably in gliomas, and various types of skin cancer suggesting a potential role in the pathogenesis of these solid malignancies. Thus, IDH1 and IDH2 could be useful as molecular therapy targets. Furthermore, patients bearing the IDH1 mutation can be benefitted from the ivosidenib or recently developed IDH1 mutant-specific peptide vaccine (IDH1-vac) and may also serve as diagnostic markers in these cancers. The IDH1 and IDH2 gene mutations can be used in clinical practice as strong prognostic biomarkers in gliomas as they could predict better survival.
This work represents an advance in biomedical science because it shows IDH1 and IDH2 mutational spectrum, significant prevalence in large cancer series and benefit of testing them for prognosis and therapeutic management.
Summary Table
What Is Known About This Subject?
• Isocitrate dehydrogenase genes, IDH1 and IDH2 have been demonstrated to be altered in brain cancers
• The IDH1 and IDH2 mutations were shown to predict the outcome of the patients with various brain malignancies
• Mutant IDH1 bearing gliomas can be therapeutically benefited from recently developed IDH1 mutant-specific peptide vaccine
What This Work Adds
• Analyses of IDH1 and IDH2 mutations in large malignant series shows a complete spectrum of these mutations in human cancers
• The IDH1 and IDH2 somatic mutations play a significant role not only in brain tumors but also in other malignancies
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
AM conceived and conducted the study, collected and analyzed the data and wrote the manuscript. AA critically reviewed the data and the manuscript. AM and AA finalized and approved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
Siegel RL Miller KD Fuchs HE Jemal A . Cancer Statistics, 2021. CA A Cancer J Clin (2021) 71:7–33. 10.3322/caac.21654
2.
Perri F Pisconti S Della Vittoria Scarpati G . P53 Mutations and Cancer: a Tight Linkage. Ann Transl Med (2016) 4:522. 10.21037/atm.2016.12.40
3.
Davies H Bignell GR Cox C Stephens P Edkins S Clegg S et al Mutations of the BRAF Gene in Human Cancer. Nature (2002) 417:949–54. 10.1038/nature00766
4.
Murugan AK Qasem E Al-Hindi H Shi Y Alzahrani AS . Classical V600E and Other Non-hotspot BRAF Mutations in Adult Differentiated Thyroid Cancer. J Transl Med (2016) 14:204. 10.1186/s12967-016-0958-x
5.
Murugan AK Grieco M Tsuchida N . RAS Mutations in Human Cancers: Roles in Precision Medicine. Semin Cancer Biol (2019) 59:23–35. 10.1016/j.semcancer.2019.06.007
6.
Samuels Y Wang Z Bardelli A Silliman N Ptak J Szabo S et al High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science (2004) 304:554. 10.1126/science.1096502
7.
Murugan A Hong N Fukui Y Munirajan A Tsuchida N . Oncogenic Mutations of the PIK3CA Gene in Head and Neck Squamous Cell Carcinomas. Int J Oncol (2008) 32:101–11. 10.3892/ijo.32.1.101
8.
Paez JG Jänne PA Lee JC Tracy S Greulich H Gabriel S et al EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science (2004) 304:1497–500. 10.1126/science.1099314
9.
Stephens P Hunter C Bignell G Edkins S Davies H Teague J et al Intragenic ERBB2 Kinase Mutations in Tumours. Nature (2004) 431:525–6. 10.1038/431525b
10.
van Haaften G Dalgliesh GL Davies H Chen L Bignell G Greenman C et al Somatic Mutations of the Histone H3K27 Demethylase Gene UTX in Human Cancer. Nat Genet (2009) 41:521–3. 10.1038/ng.349
11.
Chen Y Takita J Choi YL Kato M Ohira M Sanada M et al Oncogenic Mutations of ALK Kinase in Neuroblastoma. Nature (2008) 455:971–4. 10.1038/nature07399
12.
Murugan AK Xing M . Anaplastic Thyroid Cancers Harbor Novel Oncogenic Mutations of the ALK Gene. Cancer Res (2011) 71:4403–11. 10.1158/0008-5472.can-10-4041
13.
Liu X Bishop J Shan Y Pai S Liu D Murugan AK et al Highly Prevalent TERT Promoter Mutations in Aggressive Thyroid Cancers. Endocr Relat Cancer (2013) 20:603–10. 10.1530/erc-13-0210
14.
Murugan AK . mTOR: Role in Cancer, Metastasis and Drug Resistance. Semin Cancer Biol (2019) 59:92–111. 10.1016/j.semcancer.2019.07.003
15.
Parsons DW Jones S Zhang X Lin JC-H Leary RJ Angenendt P et al An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science (2008) 321:1807–12. 10.1126/science.1164382
16.
Yan H Parsons DW Jin G McLendon R Rasheed BA Yuan W et al IDH1andIDH2Mutations in Gliomas. N Engl J Med (2009) 360:765–73. 10.1056/nejmoa0808710
17.
Murugan AK Bojdani E Xing M . Identification and Functional Characterization of Isocitrate Dehydrogenase 1 (IDH1) Mutations in Thyroid Cancer. Biochem Biophys Res Commun (2010) 393:555–9. 10.1016/j.bbrc.2010.02.095
18.
Geisbrecht BV Gould SJ . The Human PICD Gene Encodes a Cytoplasmic and Peroxisomal NADP+-dependent Isocitrate Dehydrogenase. J Biol Chem (1999) 274:30527–33. 10.1074/jbc.274.43.30527
19.
Fathi AT Sadrzadeh H Comander AH Higgins MJ Bardia A Perry A et al Isocitrate Dehydrogenase 1 ( IDH1 ) Mutation in Breast Adenocarcinoma Is Associated with Elevated Levels of Serum and Urine 2‐Hydroxyglutarate. Oncologist (2014) 19:602–7. 10.1634/theoncologist.2013-0417
20.
Kosmider O Gelsi-Boyer V Slama L Dreyfus F Beyne-Rauzy O Quesnel B et al Mutations of IDH1 and IDH2 Genes in Early and Accelerated Phases of Myelodysplastic Syndromes and MDS/myeloproliferative Neoplasms. Leukemia (2010) 24:1094–6. 10.1038/leu.2010.52
21.
Amary MF Bacsi K Maggiani F Damato S Halai D Berisha F et al IDH1 and IDH2 Mutations Are Frequent Events in central Chondrosarcoma and central and Periosteal Chondromas but Not in Other Mesenchymal Tumours. J Pathol (2011) 224:334–43. 10.1002/path.2913
22.
Kato Kaneko M Liu X Oki H Ogasawara S Nakamura T Saidoh N et al Isocitrate Dehydrogenase Mutation Is Frequently Observed in Giant Cell Tumor of Bone. Cancer Sci (2014) 105:744–8. 10.1111/cas.12413
23.
Xu W Yang H Liu Y Yang Y Wang P Kim S-H et al Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell (2011) 19:17–30. 10.1016/j.ccr.2010.12.014
24.
Dang L White DW Gross S Bennett BD Bittinger MA Driggers EM et al Cancer-associated IDH1 Mutations Produce 2-hydroxyglutarate. Nature (2009) 462:739–44. 10.1038/nature08617
25.
Chowdhury R Yeoh KK Tian YM Hillringhaus L Bagg EA Rose NR et al The Oncometabolite 2‐hydroxyglutarate Inhibits Histone Lysine Demethylases. EMBO Rep (2011) 12:463–9. 10.1038/embor.2011.43
26.
Turcan S Rohle D Goenka A Walsh LA Fang F Yilmaz E et al IDH1 Mutation Is Sufficient to Establish the Glioma Hypermethylator Phenotype. Nature (2012) 483:479–83. 10.1038/nature10866
27.
Gao J Aksoy BA Dogrusoz U Dresdner G Gross B Sumer SO et al Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal (2013) 6:pl1. 10.1126/scisignal.2004088
28.
Wang XW Ciccarino P Rossetto M Boisselier B Marie Y Desestret V et al IDH Mutations: Genotype-Phenotype Correlation and Prognostic Impact. Biomed Res Int (2014) 2014:540236. 10.1155/2014/540236
29.
Stancheva G Goranova T Laleva M Kamenova M Mitkova A Velinov N et al IDH1/IDH2 but Not TP53 Mutations Predict Prognosis in Bulgarian Glioblastoma Patients. Biomed Res Int (2014) 2014:654727. 10.1155/2014/654727
30.
Myung JK Cho HJ Park C-K Kim S-K Phi JH Park S-H . IDH1 Mutation of Gliomas with Long-Term Survival Analysis. Oncol Rep (2012) 28:1639–44. 10.3892/or.2012.1994
31.
Juratli TA Kirsch M Geiger K Klink B Leipnitz E Pinzer T et al The Prognostic Value of IDH Mutations and MGMT Promoter Status in Secondary High-Grade Gliomas. J Neurooncol (2012) 110:325–33. 10.1007/s11060-012-0977-2
32.
Xia L Wu B Fu Z Feng F Qiao E Li Q et al Prognostic Role of IDH Mutations in Gliomas: a Meta-Analysis of 55 Observational Studies. Oncotarget (2015) 6:17354–65. 10.18632/oncotarget.4008
33.
Chen J-R Yao Y Xu H-Z Qin Z-Y . Isocitrate Dehydrogenase (IDH)1/2 Mutations as Prognostic Markers in Patients with Glioblastomas. Medicine (Baltimore) (2016) 95:e2583. 10.1097/md.0000000000002583
34.
Tabei Y Kobayashi K Saito K Shimizu S Suzuki K Sasaki N et al Survival in Patients with Glioblastoma at a First Progression Does Not Correlate with Isocitrate Dehydrogenase (IDH)1 Gene Mutation Status. J Clin Oncol (2021) 51:45–53. 10.1093/jjco/hyaa162
35.
Popovici-Muller J Lemieux RM Artin E Saunders JO Salituro FG Travins J et al Discovery of AG-120 (Ivosidenib): A First-In-Class Mutant IDH1 Inhibitor for the Treatment of IDH1 Mutant Cancers. ACS Med Chem Lett (2018) 9:300–5. 10.1021/acsmedchemlett.7b00421
36.
Angelakas A Lamarca A Hubner RA McNamara MG Valle JW . Ivosidenib: an Investigational Drug for the Treatment of Biliary Tract Cancers. Expert Opin Investig Drugs (2021) 30:301–7. 10.1080/13543784.2021.1900115
37.
Platten M Bunse L Wick A Bunse T Le Cornet L Harting I et al A Vaccine Targeting Mutant IDH1 in Newly Diagnosed Glioma. Nature (2021) 592:463–8. 10.1038/s41586-021-03363-z
Summary
Keywords
IDH1 , IDH2 , mutation, cancer, glioma, dehydrogenase, isocitrate
Citation
Murugan AK and Alzahrani AS (2022) Isocitrate Dehydrogenase IDH1 and IDH2 Mutations in Human Cancer: Prognostic Implications for Gliomas. Br J Biomed Sci 79:10208. doi: 10.3389/bjbs.2021.10208
Received
10 November 2021
Accepted
02 December 2021
Published
31 January 2022
Volume
79 - 2021
Updates
Copyright
© 2022 Murugan and Alzahrani.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. K. Murugan, akmurugan@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.