Abstract
Cryptosporidium, the most frequently reported parasite in Scotland, causes gastrointestinal illness resulting in diarrhoea, nausea and cramps. Two species are responsible for most cases: Cryptosporidium hominis (C. hominis) and Cryptosporidium parvum (C. parvum). Transmission occurs faecal-orally, through ingestion of contaminated food and water, or direct contact with faeces. In 2020, the COVID-19 pandemic led to global restrictions, including national lockdowns to limit viral transmission. Such interventions led to decreased social mixing, and reduced/no local and international travel, which are factors associated with transmission of multiple communicable diseases, including cryptosporidiosis. This report assessed the impact of the pandemic on Scottish cryptosporidiosis cases, and identified changes in circulating molecular variants of Cryptosporidium species. Molecular data generated using real time PCR and GP60 nested-PCR assays on laboratory-confirmed cryptosporidiosis cases reported during 2018–22 were analysed. The Scottish Microbiology Reference Laboratories (SMiRL), Glasgow, received 774 Cryptosporidium-positive faeces during 2018–22, of which 486 samples were successfully subtyped. During this time period, C. hominis (n = 155; 21%) and C. parvum (n = 572; 77%) were the most commonly detected species. The total number of cases during 2020, which was greatly affected by the pandemic, was markedly lower in comparison to case numbers in the 2 years before and after 2020. The most predominant C. hominis family detected prior to 2020 was the Ib family which shifted to the Ie family during 2022. The most common C. parvum variant during 2018–22 was the IIa family, however a rise in the IId family was observed (n = 6 in 2018 to n = 25 in 2022). The dominant C. hominis subtype IbA10G2, which accounted for 71% of C. hominis subtypes in 2018–19 was superseded by three rare subtypes: IeA11G3T3 (n = 15), IdA16 (n = 8) and IbA9G3 (n = 3) by 2022. Frequently reported C. parvum subtypes in 2018–19 were IIaA15G2R1 and IIaA17G1R1, accounting for 59% of total C. parvum subtypes. By 2022, IIaA15G2R1 remained the most common (n = 28), however three unusual subtypes in Scotland emerged: IIdA24G1 (n = 7), IIaA16G3R1 (n = 7) and IIaA15G1R2 (n = 7). Continuous monitoring of Cryptosporidium variants following the pandemic will be essential to explore further changes and emergence of strains with altered virulence.
Introduction
Cryptosporidium is the most common parasitic aetiological agent of gastrointestinal infections reported in Scotland, United Kingdom [1]. The infective stage, known as the oocyst, is transmitted through ingestion of contaminated food or water, or by direct contact with infected faeces from human and/or animal sources [2]. Infection results in symptoms including diarrhoea, nausea, cramps, fatigue and weight loss, which are often self-limiting in the immunocompetent, however can be chronic and potentially life-threatening in the immunocompromised and young children [2]. Throughout the UK, the majority of human cryptosporidiosis cases are caused by two Cryptosporidium species; the anthroponotic Cryptosporidium hominis (C. hominis), and the zoonotic Cryptosporidium parvum (C. parvum) [3]. The oocysts of C. parvum and C. hominis appear morphologically identical by microscopy, therefore molecular-based detection methods are required to fully speciate this parasite [1, 4]. Over the past few decades, the ability to determine molecular variants of Cryptosporidium at a genotypic level has become possible using sophisticated laboratory tools. Most recently, a multi-locus variable number of tandem repeats analysis (MLVA) scheme has been developed of high discriminatory power which has yet to be implemented globally [5]. The majority of specialist laboratories currently amplify a parasite-specific 60 kDa glycoprotein gene (GP60), to determine the barcode of this hypervariable microsatellite region when subtyping Cryptosporidium [6]. Possessing the ability to subtype this parasite has proven to be indispensable for monitoring circulating molecular variants within communities, and managing local, national and international outbreaks.
The onset of the Coronavirus disease, COVID-19 pandemic caused by the SARS-CoV-2 virus, resulted in sudden, major change to everyday life on a global scale, with the first confirmed COVID-19 case in Scotland occurring on 1st March 2020 [7]. The UK and Scottish Governments applied measures most notably, regional and national lockdowns, in a bid to impede viral transmission. By 29th May 2020, Scotland began its route map out of lockdown, which over time, permitted pre-pandemic activities to gradually recommence. However further shorter lockdowns were implemented both at a regional and national level in response to rising COVID-19 levels [7]. Implementation of such measures resulted in decreased social mixing, closure of non-essential businesses such as food and drink venues, a ban on international travel, and improved hand hygiene, all of which had the potential to impact on the number of cases, and molecular subtypes of Cryptosporidium within the population.
This report highlights the changing dynamics of Cryptosporidium at a molecular level from 2018–22 in Scotland to gain a better understanding of the impact of COVID-19 restrictions and human cryptosporidiosis.
Materials and Methods
During 2018–22, a total of 2063 cases of laboratory-confirmed cryptosporidiosis were reported to Public Health Scotland (PHS) (5-year average n = 611; range 502–786 cases; personal communication). Of those laboratory-confirmed cases, a total of 744 faecal samples positive for Cryptosporidium oocysts, antigen or DNA were received at the Scottish Microbiology Reference Laboratories (SMiRL) Glasgow, UK for molecular analysis. Faeces were sent from National Health Service (NHS) Diagnostic Microbiology Laboratories across Scotland as part of the Scottish Government-funded Cryptosporidium Surveillance and Outbreak Service. As funding was not available to subtype every positive sample, a selection of faeces from rural, semi-rural and urban areas were subtyped to give a useful indication of circulating types within Scotland. Multiple samples received from the same case within a 4-week period were classed as one sample.
Samples were transported and stored at room temperature. On arrival, faeces were concentrated using Parasep concentrators (Apicor, Berkshire, United Kingdom), and Cryptosporidium DNA extracted on the automated NucliSENS EasyMag platform (BioMerieux, Basingstoke, United Kingdom). To determine if samples were C. hominis or C. parvum, a previously described real-time PCR assay was employed using the Light Cycler® 480 platform (Roche Diagnostics, West Sussex, United Kingdom) [8]. The molecular profile of each sample was assigned by amplifying a region of the GP60 gene followed by bi-directional sequencing on the Applied Biosystems 3500XL (ThermoFisher Scientific, Inchinnan, United Kingdom), as previously described [1]. Sequences were aligned to reference databases (EMBI and NCBI BLASTn) and repeat sequences determined manually.
Results
Of the 744 faecal DNA samples speciated, the majority were found to be either C. hominis (n = 155; 21%) or C. parvum (n = 572; 77%). Other species accounted for less than 3% of the total (C. cuniculus n = 5; C. ubiquitum n = 6; C. meleagridis n = 2; C. canis n = 1; C. erinacei n = 1; mixed infection with C. parvum and C. hominis n = 2).
For C. hominis, a total of 54 cases were identified in 2018 and a further 54 cases in 2019, with the expected late summer/autumn rise observed in September (Figure 1A). In 2020 and 2021 which were greatly impacted by lockdowns and restrictions, only five cases were identified with no autumn peak evident. During 2022, when COVID-19 lockdowns and restrictions had ended, the total number of C. hominis cases (n = 42) were similar to pre-pandemic levels (Figure 1A).
FIGURE 1
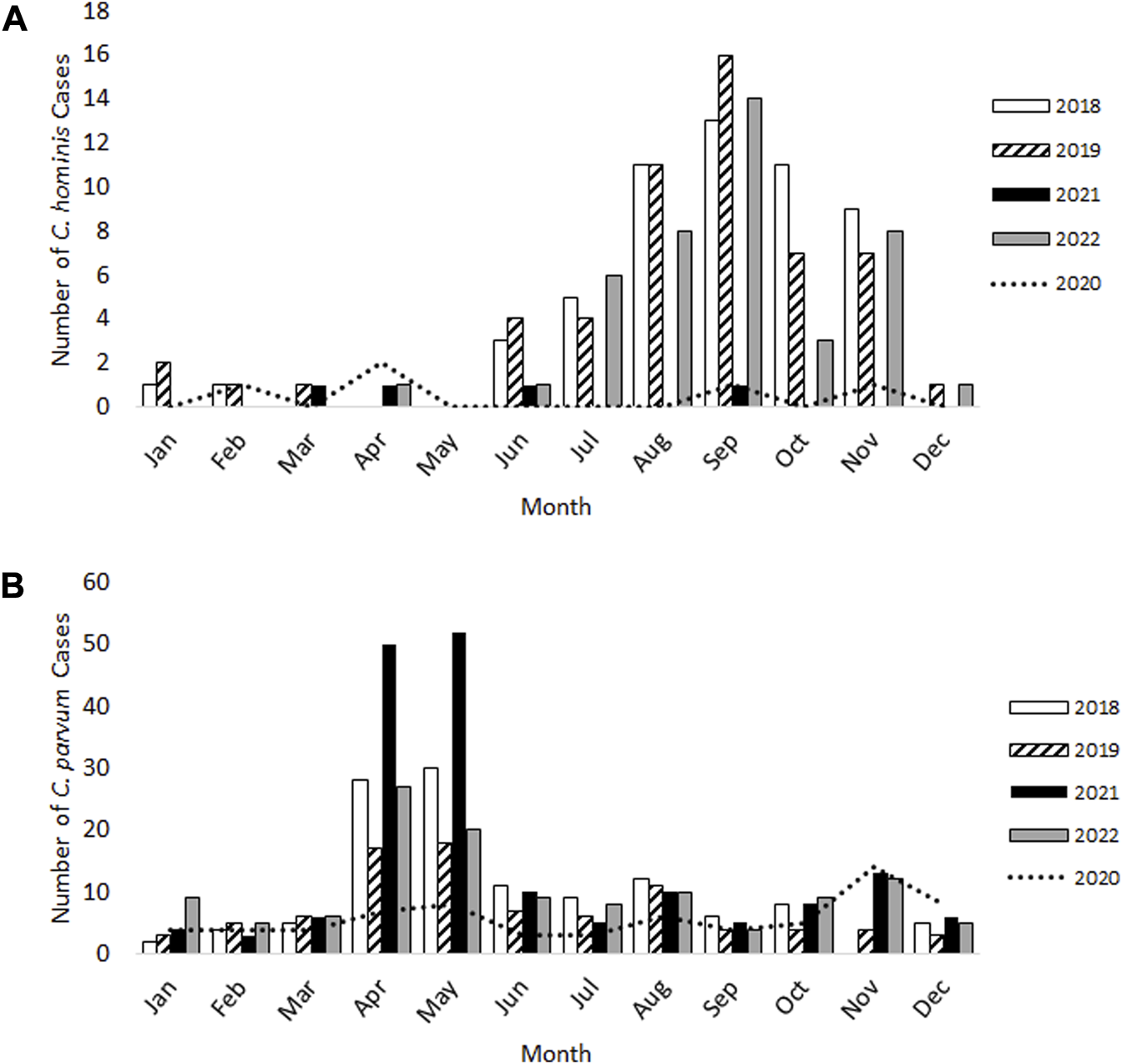
Laboratory-confirmed cases of Cryptosporidium hominis(A) and Cryptosporidium parvum(B) in Scotland from 2018–22.
For C. parvum, 120 cases and 88 cases were reported during 2018 and 2019 respectively, with the expected rise occurring over springtime (April-May) and again in August (Figure 1B). Whilst fewer C. parvum cases were reported in 2020 (n = 70), an additional, small peak was observed in November 2020 (n = 14 cases), which had not occurred in the previous 2 years (Figure 1B). This increase was also evident during 2021 and 2022 (Figure 1B). In 2021, the total number of C. parvum cases was 172, where the expected springtime increase during April-May was observed, but the number of cases were 1.8-fold and 2.8-fold greater than those identified in the same months during 2018 and 2019, respectively. In 2022, 124 C. parvum cases were reported, and the spring peak occurred, similar to 2018 and 2019 (Figure 1B).
Of the 744 faecal DNA samples speciated, 486 (65%) were successfully subtyped. Sixty-eight (14%) were C. hominis subtypes, and 415 (85%) were C. parvum subtypes. A further three samples identified as C. cuniculus were successfully subtyped (data not shown).
There was a shift in the most predominant C. hominis subtype family where the Ib family was most frequently reported in 2018 (n = 29), with only one case each of the Ie and Id variants. However, by the end of 2022, only three cases of the Ib family were identified, compared to nine cases of the Id, and 15 of the Ie families (Table 1).
TABLE 1
| C. hominis | C. parvum | |||||
|---|---|---|---|---|---|---|
| Year | Ib | Id | Ie | Ila | Ilc | Ild |
| 2018 | 29 | 1 | 1 | 62 | 1 | 6 |
| 2019 | 2 | 0 | 0 | 56 | 2 | 11 |
| 2020 | 1 | 0 | 0 | 30 | 2 | 19 |
| 2021 | 0 | 0 | 1 | 116 | 1 | 17 |
| 2022 | 3 | 9 | 15 | 65 | 2 | 25 |
Most commonly detected Cryptosporidium hominis and Cryptosporidium parvum molecular families from 2018–22.
Before the implementation of 2020 COVID-19 lockdowns and restrictions, a total of 25 cases of the common C. hominis IbA10G2 subtype were identified in 2018–19 (71% of total C. hominis subtyped in 2018–19). In 2020, only two cases of C. hominis were subtyped (IbA10G2 n = 1, IaA24R2 n = 1). During 2021–22, there were no reports of the common IbA10G2 subtype but the emergence of three rare subtypes in Scotland were documented; IeA11G3T3 (n = 15); IdA16 (n = 8) and IbA9G3 (n = 3) (Table 2). Of the 415 C. parvum samples successfully subtyped, the three most common molecular families were the IIa (n = 329), IId (n = 78) and the IIc (n = 8) (Table 1). Whilst the IIa family remained the most common family throughout 2018–22, of interest, there was a gradual increase in numbers of the IId family (Table 1). Of note, for C. parvum, the total number of different subtypes identified in the springtime peak (April-May) throughout 2018–22 varied. During 2018–2020, there was an average of eight different subtypes. However in 2021, the number of different subtypes increased to 18, and remained elevated in 2022 with 13 different subtypes identified.
TABLE 2
| Year | ||||||
|---|---|---|---|---|---|---|
| 2018 | 2019 | 2020 | 2021 | 2022 | ||
| C. hominis | IbA10G2 | 23 | 2 | 1 | 0 | 0 |
| IeA11G3T3 | 1 | 0 | 0 | 1 | 15 | |
| IbA12G3 | 5 | 0 | 0 | 0 | 0 | |
| IaA24R2 | 0 | 0 | 1 | 0 | 0 | |
| IdA16 | 0 | 0 | 0 | 0 | 8 | |
| IbA9G3 | 0 | 0 | 0 | 0 | 3 | |
| C. parvum | IIaA16G2R1 | 0 | 1 | 0 | 1 | 1 |
| IIaA17G1R1 | 20 | 13 | 5 | 24 | 6 | |
| IIaA15G2R1 | 30 | 19 | 15 | 54 | 28 | |
| IIdA24G1 | 0 | 2 | 2 | 0 | 7 | |
| IIaA18G2R1 | 2 | 1 | 1 | 3 | 0 | |
| IIcA5G3 | 1 | 2 | 2 | 1 | 2 | |
| IIaA13G1R2 | 0 | 1 | 0 | 5 | 1 | |
| IIdA17G1 | 3 | 0 | 4 | 5 | 5 | |
| IIaA21G4R1 | 0 | 5 | 1 | 0 | 0 | |
| IIdA22G1 | 1 | 0 | 6 | 3 | 3 | |
| IIaA19G1R1 | 1 | 0 | 0 | 5 | 0 | |
| IIaA15G1R2 | 2 | 3 | 2 | 1 | 7 | |
| IIaA16G3R1 | 1 | 1 | 1 | 1 | 7 | |
| IIdA19G1 | 1 | 1 | 4 | 3 | 5 | |
Cryptosporidium hominis and Cryptosporidium parvum subtypes detected from 2018–22.
The most common subtypes in 2018–19 were IIaA15G2R1 (n = 30 and n = 19, 2018 and 2019, respectively) and IIaA17G1R1 (n = 20 and n = 13 in 2018 and 2019, respectively), which accounted for 59% of cases subtyped. In 2020, whilst IIaA15G2R1 remained the most frequently reported subtype (n = 15; 29% of all C. parvum subtypes), the second most common was a IId subtype, namely, IIdA22G1 (n = 6), which accounted for 12% of all C. parvum isolates typed that year. In 2021, the IIaA15G2R1 and IIaA17G1R1 subtypes were most frequently observed (n = 54 and n = 24, respectively), accounting for 40% and 18% of the total. However, during 2022, the IIaA15G2R1 remained the most common (n = 28; 30%), but numbers of more unusual subtypes IIdA24G1 (n = 7), IIaA16G3R1 (n = 7) and IIaA15G1R2 (n = 7) were similar to the common IIaA17G1R1 type (n = 6) (Table 2).
Discussion
In this report, we highlight that during the past 5 years, C. hominis only represented around one-fifth of those cases that were successfully typed, with C. parvum accounting for almost 80%. This is in contrast to previously published data from Scotland during 2012–13 and unpublished data from 2014–17 (data not shown), where similar numbers of C. hominis and C. parvum cases were reported [1]. This change in recent years is likely to reflect the altered human behaviours as a result of the pandemic restrictions, which included improved hand hygiene, reduced social interactions, a reduction in dining out at food and drink establishments, and reduced or no local/international travel [7, 9].
Similar to published data [1], this study found that human cryptosporidiosis cases normally occurred seasonally, with a peak in late-Spring of mainly zoonotic C. parvum infections coinciding with the lambing/calving season and open farm outbreaks, and a second peak occurring in late-Summer/early-Autumn mainly comprising of C. hominis infections assumed to be associated with an increase in human to human transmission locally, and also international travel [10].
Further, more in-depth analysis of epidemiological data for each case is necessary to ascertain what exposure(s) were likely to have resulted in each infection, but with the demand for limited resources within Public Health Scotland to support and prioritise COVID investigations, this has not been possible in recent times.
During 2020, which was greatly impacted by restrictions and lockdowns implemented to limit transmission of COVID-19, cases of cryptosporidiosis were markedly lower which is a similar pattern for the incidence of other gastrointestinal (GI) illnesses in the United Kingdom [11, 12]. A recent report on the effects of COVID-19 restrictions on cryptosporidiosis cases in England and Wales described similar findings to that observed in Scotland, where the incidence of C. hominis and C. parvum decreased markedly from the onset of COVID-19 restrictions [13]. These finding differed to that of a study from New Zealand, which reported an absence of C. hominis cases within the first few months of COVID-19 restrictions, yet C. parvum cases continued to be reported at a similar level [14].
Indirect effects of the national lockdown and imposed COVID-19 restrictions are likely to have impacted on the observed decrease in cryptosporidiosis cases during 2020. The closure of all non-essential businesses, including those previously linked to cryptosporidiosis outbreaks, such as swimming pools, open farms, food outlets and restaurants, may have contributed to lower cases being observed during this time [9, 12, 15]. Improved hand hygiene was encouraged throughout the pandemic, which is also an important measure in minimising transmission of GI infections, and is therefore likely to have decreased human-to-human transmission [16]. Starting 29th May 2020, the route map out of lockdown commenced in Scotland, which gradually permitted more socialising and pre-pandemic activities. However during this period of returning to normality, regional lockdowns were implemented in areas of Scotland identified to have high numbers of COVID-19 cases [7]. On implementation of the route map out of lockdown, there was a general reluctance to travel which is likely to have impacted on the number of cases, especially C. hominis during late-Summer/early-Autumn of 2020 [17]. Perhaps the potential loss in confidence to book holidays for fear of cancellations, and the need to be fully vaccinated when travelling to certain areas, impacted on numbers not fully returning to pre-pandemic levels by 2022 [18]. Whilst this study reported a decrease in laboratory-confirmed cryptosporidiosis cases during 2020, the true number of cases within the community is likely to be under-represented as a result of the public being advised to only contact healthcare providers in emergencies. Although this alleviated pressures on General Practitioners (GPs) and hospitals during the pandemic, it is likely to have resulted in individuals being reluctant to contact GPs with gastrointestinal illness.
The large spike in C. parvum cases observed in April and May of 2021, is likely to reflect the reopening of non-essential businesses including swimming pools, food outlets, petting farms/open farms, which enabled mixing of groups of individuals, and increased exposure to infected humans and animals, especially direct contact with young animals.
Over the time period covered in this study, the most predominant C. hominis family detected changed from the Ib family, which is the most common C. hominis family in Europe [19], to the Ie family followed by the Id family, both of which are commonly found in low/middle income countries [19]. The three rare C. hominis subtypes identified, namely, IeA11G3T3, IdA16 and IbA9G3 were likely to have been imported to Scotland once international travel began after the easing of pandemic lockdowns. One of the three rare C. hominis variants, namely, IeA11G3T3, is associated with infection in Asia, Africa and South America, and was previously identified in Scotland, with epidemiological links to a contaminated swimming pool [1, 20–22]. Subtype IdA16 has been previously identified in England, with most recent infections associated with a swimming pool outbreak [23]. To our knowledge, this is the first account of the IdA16 subtype within the Scottish population. A recent study found IdA16 infections identified in Sweden were associated with travel from Sri Lanka and China [24]. Lastly, the rare IbA9G3 subtype has been reported from Malawi, Kenya and India [19]. It has been previously reported that detection of rare C. hominis subtypes is associated with international travel [25]. Therefore, detection of these rare C. hominis subtypes post-2020 within the Scottish population was likely associated with increased travel beyond Europe following the pandemic, although as international travel had not yet reach pre-pandemic levels, consideration for possible imported contaminated consumables should be given [18].
In this report, the most predominant C. parvum family was IIa, in particular, IIaA15G2R1 which has also been reported from calves and geese in Scotland [26, 27]. Of interest, there was an increase in the number of IId family cases which is found throughout Europe, and is associated with infection of ruminants, including sheep, goats and calves [19, 27]. Throughout the pandemic, an increase in outdoor exercise and leisure activities e.g., wild water swimming was observed. Spending more time in green spaces is likely to have resulted in greater exposure to farmed animals and/or their faeces, resulting in an increase in the incidence of infection with the IId family [28, 29]. This may have also contributed to the increase in number of different C. parvum subtypes observed between April-May in 2021 and 2022. One of the more unusual C. parvum subtypes commonly observed post-pandemic namely, IIdA24G1, was previously identified in European hedgehogs (Erinaceus europaeus), Spanish lambs and humans in Sweden and Australia [30–33]. Although of interest, to our knowledge, the IId family has not been identified from animals within Scotland (Personal communications, Moredun Institute, Scotland). There is the possibility that the emergence of the IId family in Scotland is via an imported consumable as certain variants including the IIdA24G1 subtype have been associated with European outbreaks involving salad items [34]. The rare IIaA16G3R1, has previously been associated with cattle infections [19], and IIaA15G1R2, has been reported from UK outbreaks involving open farms [35]. As only a proportion of Cryptosporidium-positive faeces are typed in Scotland, due to the limited funds available, combined with the presence of PCR inhibitors in faeces, only 44% of C. hominis positive samples and 73% of C. parvum positive samples were subtyped.
This work represents an advance in biomedical science because it highlights that Cryptosporidium continues to be a significant cause of gastrointestinal disease, and the COVID-19 pandemic has resulted in changes to both the dominant species and subtypes circulating within the Scottish population. It will be crucial to monitor the molecular variants over the coming years to see if these changes remain stable, and if new and emerging strains appear with altered virulence as we exist in the post-pandemic era.
Statements
Data availability statement
The datasets presented in this article are not readily available because Public Health Scotland are owners of the data. Enquires to access the data should be made to the corresponding author. Requests to access the datasets should be directed to ross.bacchetti@ggc.scot.nhs.uk.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardian/next of kin in accordance with the local legislation and institutional requirements.
Author contributions
RB contributed towards data analysis and manuscript preparation. LC contributed to sample processing. LB contributed to data collection. CA contributed towards manuscript preparation. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank laboratory staff within SMiRL, Glasgow for processing samples and PHS colleagues for data information.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
Deshpande AP Jones BL Connelly L Pollock KG Brownlie S Alexander CL . Molecular Characterization of Cryptosporidium Parvum Isolates From Human Cryptosporidiosis Cases in Scotland. Parasitology (2015) 142(2):318–25. 10.1017/S0031182014001346
2.
Bouzid M Hunter PR Chalmers RM Tyler KM . Cryptosporidium Pathogenicity and Virulence. Clin Microbiol Rev (2013) 26(1):115–34. 10.1128/CMR.00076-12
3.
Putignani L Menichella D . Global Distribution, Public Health and Clinical Impact of the Protozoan Pathogen Cryptosporidium. Interdiscip Perspect Infect Dis (2010) 2010:753512. 10.1155/2010/753512
4.
Hønsvall BK Robertson LJ . Real-Time Nucleic Acid Sequence-Based Amplification (NASBA) Assay Targeting MIC1 for Detection of Cryptosporidium Parvum and Cryptosporidium Hominis Oocysts. Exp Parasitol (2017) 172:61–7. 10.1016/j.exppara.2016.12.009
5.
Robinson G Pérez-Cordón G Hamilton C Katzer F Connelly L Alexander CL et al Validation of a Multilocus Genotyping Scheme for Subtyping Cryptosporidium Parvum for Epidemiological Purposes. Food Waterborne Parasitol (2022) 27:e00151. 10.1016/j.fawpar.2022.e00151
6.
Strong WB Gut J Nelson RG . Cloning and Sequence Analysis of a Highly Polymorphic Cryptosporidium Parvum Gene Encoding a 60-Kilodalton Glycoprotein and Characterization of its 15- and 45-Kilodalton Zoite Surface Antigen Products. Infect Immun (2000) 68(7):4117–34. 10.1128/IAI.68.7.4117-4134.2000
7.
SPICe Spotlight. Timeline of Coronavirus (COVID-19) in Scotland – SPICe Spotlight | Solas Air SPICe (2023). [Internet]. Available from: https://spice-spotlight.scot/2022/12/16/timeline-of-coronavirus-covid-19-in-scotland/ (Accessed March 24, 2023).
8.
Hadfield SJ Robinson G Elwin K Chalmers RM . Detection and Differentiation of Cryptosporidium Spp. In Human Clinical Samples by Use of Real-Time PCR. J Clin Microbiol (2011) 49(3):918–24. 10.1128/JCM.01733-10
9.
Love NK Elliot AJ Chalmers RM Douglas A Gharbia S McCormick J et al Impact of the COVID-19 Pandemic on Gastrointestinal Infection Trends in England, February-July 2020. BMJ Open (2022) 12(3):e050469. 10.1136/bmjopen-2021-050469
10.
Cacciò SM Chalmers RM . Human Cryptosporidiosis in Europe. Clin Microbiol Infect (2016) 22(6):471–80. 10.1016/j.cmi.2016.04.021
11.
Public Health Scotland. Gastrointestinal and Zoonoses - Biennial Report 2020 to 2021 - Gastrointestinal and Zoonoses - Publications - Public Health Scotland (2023). [Internet]. Available from: https://www.publichealthscotland.scot/publications/gastrointestinal-and-zoonoses/gastrointestinal-and-zoonoses-biennial-report-2020-to-2021/ (Accessed April 3, 2023).
12.
Chalmers RM McCarthy N Barlow KL Stiff R . An Evaluation of Health Protection Practices for the Investigation and Management of Cryptosporidium in England and Wales. J Public Health (Oxf) (2018) 40(1):114–20. 10.1093/pubmed/fdw143
13.
Adamson JP Chalmers RM Thomas DR Elwin K Robinson G Barrasa A . Impact of the COVID-19 Restrictions on the Epidemiology of Cryptosporidium Spp. In England and Wales, 2015-2021: A Time Series Analysis. J Med Microbiol (2023) 72(6). 10.1099/jmm.0.001693
14.
Knox MA Garcia -RJC Ogbuigwe P Pita A Velathanthiri N Hayman DTS . Absence of Cryptosporidium Hominis and Dominance of Zoonotic Cryptosporidium Species in Patients After COVID-19 Restrictions in Auckland, New Zealand. Parasitology (2021) 148(11):1288–92. 10.1017/S0031182021000974
15.
Polubotho P Denvir L Connelly L Anderson E Alexander CL . The First UK Report of a Rare Cryptosporidium Hominis Genetic Variant Isolated During a Complex Scottish Swimming Pool Outbreak. J Med Microbiol (2021) 70(3):001289. 10.1099/jmm.0.001289
16.
UK Government. Gastrointestinal Infections: Guidance for Public Health Management - GOV.UK (2020). [Internet] Available from: https://www.gov.uk/government/publications/gastrointestinal-infections-guidance-for-public-health-management (Accessed March 17, 2023).
17.
Office of National Statistics. Overseas Travel and Tourism - Office for National Statistics (2021). [Internet]. Available from: https://www.ons.gov.uk/peoplepopulationandcommunity/leisureandtourism/articles/overseastravelandtourism2020/2021-05-24 (Accessed March 17, 2023).
18.
Office of National Statistics. Overseas Travel and Tourism - Office for National Statistics (2023). [Internet]. Available from: https://www.ons.gov.uk/peoplepopulationandcommunity/leisureandtourism/bulletins/overseastravelandtourism/octoberandnovember2022provisionalresults (Accessed March 17, 2023).
19.
Xiao L . Molecular Epidemiology of Cryptosporidiosis: An Update. Exp Parasitol (2010) 124(1):80–9. 10.1016/j.exppara.2009.03.018
20.
Saramago Peralta RH Néstor Velásquez J de Souza Cunha F Laura Pantano M Campos Sodré F da Silva S et al Genetic Diversity of Cryptosporidium Identified in Clinical Samples From Cities in Brazil and Argentina. Mem Inst Oswaldo Cruz (2016) 111(1):30–6. 10.1590/0074-02760150303
21.
Sharma P Sharma A Sehgal R Malla N Khurana S . Genetic Diversity of Cryptosporidium Isolates From Patients in North India. Int J Infect Dis (2013) 17(8):e601–e605. 10.1016/j.ijid.2012.12.003
22.
Adamu H Petros B Zhang G Kassa H Amer S Ye J et al Distribution and Clinical Manifestations of Cryptosporidium Species and Subtypes in HIV/AIDS Patients in Ethiopia. Plos Negl Trop Dis (2014) 8(4):e2831. 10.1371/journal.pntd.0002831
23.
Bagnall H Chalmers RM Henderson M Sorrell S Robinson G Elwin K et al Swimming Pool-Related Outbreak of a Rare Gp60 Subtype of Cryptosporidium Hominis, England, October 2016. Water (Switzerland) (2021) 13(22):3152. 10.3390/w13223152
24.
Lebbad M Winiecka-Krusnell J Stensvold CR Beser J . High Diversity of Cryptosporidium Species and Subtypes Identified in Cryptosporidiosis Acquired in Sweden and Abroad. Pathogens (2021) 10(5):523. 10.3390/pathogens10050523
25.
Chalmers RM Hadfield SJ Jackson CJ Elwin K Xiao L Hunter P . Geographic Linkage and Variation in Cryptosporidium Hominis. Emerg Infect Dis (2008) 14(3):496–8. 10.3201/eid1403.071320
26.
Wells B Paton C Bacchetti R Shaw H Stewart W Plowman J et al Cryptosporidium Prevalence in Calves and Geese Co-Grazing on Four Livestock Farms Surrounding Two Reservoirs Supplying Public Water to Mainland Orkney, Scotland. Scotland Microorg (2019) 7(11):513. 10.3390/microorganisms7110513
27.
Mammeri M Chevillot A Chenafi I Julien C Vallée I Polack B et al Molecular Characterization of Cryptosporidium Isolates From Diarrheal Dairy Calves in France. Vet Parasitol Reg Stud Reports (2019) 18:100323. 10.1016/j.vprsr.2019.100323
28.
Day BH . The Value of Greenspace Under Pandemic Lockdown. Environ Resour Econ (2020) 76(4):1161–85. 10.1007/s10640-020-00489-y
29.
Ding D Del Pozo Cruz B Green MA Bauman AE . Is the COVID-19 Lockdown Nudging People to Be More Active: A Big Data Analysis. Br J Sports Med (2020) 54(20):1183–4. 10.1136/bjsports-2020-102575
30.
Sangster L Blake DP Robinson G Hopkins TC Sa RCC Cunningham AA et al Detection and Molecular Characterisation of Cryptosporidium Parvum in British European Hedgehogs (Erinaceus Europaeus). Vet Parasitol (2016) 217:39–44. 10.1016/j.vetpar.2015.12.006
31.
Quílez J Torres E Chalmers RM Hadfield SJ Del Cacho E Sánchez-Acedo C . Cryptosporidium Genotypes and Subtypes in Lambs and Goat Kids in Spain. Appl Environ Microbiol (2008) 74(19):6026–31. 10.1128/AEM.00606-08
32.
Waldron LS Ferrari BC Power ML . Glycoprotein 60 Diversity in C. Hominis and C. Parvum Causing Human Cryptosporidiosis in NSW, Australia. Exp Parasitol (2009) 122(2):124–7. 10.1016/j.exppara.2009.02.006
33.
Gherasim A Lebbad M Insulander M Decraene V Kling A Hjertqvist M et al Two Geographically Separated Food-Borne Outbreaks in Sweden Linked by an Unusual Cryptosporidium Parvum Subtype, October 2010. Euro Surveill (2012) 17(46):20318–8. 10.2807/ese.17.46.20318-en
34.
Zahedi A Ryan U . Cryptosporidium - An Update With an Emphasis on Foodborne and Waterborne Transmission. Res Vet Sci (2020) 132:500–12. 10.1016/j.rvsc.2020.08.002
35.
Chalmers RM Robinson G Elwin K Elson R . Analysis of the Cryptosporidium Spp. And Gp60 Subtypes Linked to Human Outbreaks of Cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors (2019) 12(1):95. 10.1186/s13071-019-3354-6
Summary
Keywords
human cryptosporidiosis, molecular variants, COVID-19 pandemic, Scotland, Cryptosporidium
Citation
Bacchetti R, Connelly L, Browning L and Alexander CL. (2023) Changing Molecular Profiles of Human Cryptosporidiosis Cases in Scotland as a Result of the Coronavirus Disease, COVID-19 Pandemic. Br J Biomed Sci 80:11462. doi: 10.3389/bjbs.2023.11462
Received
11 April 2023
Accepted
02 August 2023
Published
28 August 2023
Volume
80 - 2023
Updates
Copyright
© 2023 Bacchetti, Connelly, Browning and Alexander.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ross Bacchetti, ross.bacchetti@ggc.scot.nhs.uk
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.