Abstract
Burkholderia cenocepacia is an opportunistic pathogen that is primarily associated with severe respiratory infections in people with cystic fibrosis. These bacteria have significant intrinsic resistance to antimicrobial therapy, and there is a need for more effective treatments. Bacterial zinc uptake and homeostasis systems are attractive targets for new drugs, yet our understanding of how bacteria acquire and utilise zinc remains incomplete. Here we have used RNA-sequencing and differential gene expression analysis to investigate how B. cenocepacia H111 is able to survive in zinc poor environments, such as those expected to be encountered within the host. The data shows that 201 genes are significantly differentially expressed when zinc supply is severely limited. Included in the 85 upregulated genes, are genes encoding a putative ZnuABC high affinity zinc importer, two TonB-dependent outer membrane receptors that may facilitate zinc uptake across the outer cell membrane, and a COG0523 family zinc metallochaperone. Amongst the 116 downregulated genes, are several zinc-dependent enzymes suggesting a mechanism of zinc sparring to reduce the cells demand for zinc when bioavailability is low.
Introduction
The Burkholderia cepacia complex (Bcc) is a group of at least 24 species of bacteria widely distributed in the environment [1]. They first came to prominence as opportunistic pathogens in the 1980s, causing severe respiratory infections in people with cystic fibrosis (CF) [2]. Whilst all species within the Bcc have been identified in sputum from CF patients, B. cenocepacia and B. multivorans are responsible for most infections, accounting for 85%–97% of cases [3]. Bcc infections in CF patients have a variable clinical course, although the prognosis is poor. Infections often result in an accelerated decline in pulmonary function and increased morbidity and mortality. Bcc bacteria can also lead to the development of “cepacia syndrome” that is characterised by a fatal necrotising pneumonia and bacteraemia [4]. Bcc bacteria are naturally resistant to major classes of antibiotics in clinical use, making infections particularly difficult to eradicate [5]. There are currently no standardised drug regimens/guidelines for the treatment of these infections and no vaccines available [6]. The introduction of strict infection control measures and the segregation of patients has resulted in a marked reduction in patient-to-patient transmissions in recent decades [7]. The epidemiology of Bcc bacteria is not limited to CF, with infections also occurring in people with chronic granulomatous disease [8], and regular reports of nosocomial infections in both immune compromised and immune competent individuals, often from contaminated medical supplies [9].
To survive, pathogens must be able to acquire the essential nutrients they need for growth in competition with the host. Zinc is an essential nutrient for bacteria, functioning as a catalytic cofactor or structural component of numerous proteins [10]. It is involved in fundamental cellular processes including DNA replication, regulation of gene expression and protection against oxidative damage. In some bacteria, zinc also plays a direct role in antibiotic resistance, with the production of zinc-dependent β-lactamases [11]. Overall, 5%–6% of the proteins encoded in any bacterial genome are thought to bind zinc [10]. However, since free zinc can also be toxic when present in excess, cellular zinc levels are carefully controlled by a combination of import, efflux, and storage [12]. During infection, the host attempts to deprive bacteria of the essential zinc they need to grow in a process called nutritional immunity [13]. A key aim of this study was to determine how B. cenocepacia can acquire zinc and survive in environments where zinc supply is severely restricted.
Almost all bacteria studied thus far possess ATP-binding cassette (ABC) transporters (ZnuABC) for zinc uptake in zinc poor environments [14]. These tripartite systems consist of a Zn2+ binding periplasmic protein (ZnuA), a membrane spanning permease (ZnuB) that facilitates Zn2+ transport across the plasma membrane, and a cytosolic ATPase (ZnuC) that drives transport [14]. Expression of znuABC is dependent upon intracellular zinc status and is regulated by the transcriptional repressor protein Zur [14]. In its fully Zn2+ bound state, Zur binds to specific DNA sequences (Zur boxes), within the operator regions of regulated genes. This prevents the transcription of downstream genes including znuABC avoiding the accumulation of toxic levels of zinc inside the cell [12, 14]. Some bacterial species have an additional Zur regulated protein called ZinT that acts in concert with ZnuA in the recruitment of Zn2+ within the periplasm [15].
A few bacterial species have been discovered to have additional transporters for zinc uptake across the plasma membrane. Haemophilus influenzae encodes at least two high affinity zinc uptake systems, (ZnuABC and a novel ZevAB system) [16], whilst Escherichia coli and Salmonella enterica both have a constitutively expressed low affinity ZIP-family transporter (ZupT) that import zinc under metal replete conditions [17].
It is largely assumed that passive diffusion of zinc through unspecific channels in the outer cell membrane of Gram-negative bacteria occurs in replete environments. However, this is unlikely to meet cellular demand when zinc supply is severely restricted. One potential strategy that bacteria might use to compete with the host for zinc is to secrete small molecules called “zincophores” that are like siderophores used for iron acquisition. To date, very few bacterial “zincophores” have been identified. A broad-spectrum nicotinamine-like metallophore (staphylopine) is produced by Staphylococcus aureus and is involved in the uptake of several metals, including zinc [18, 19]. In Yersinia pestis, there is evidence for the involvement of the siderophore yersiniabactin in zinc uptake [20] and in Pseudomonas aeruginosa a bona fide “zincophore” (pseudopaline) has been discovered that is regulated by Zur [21]. Several Gram-negative bacteria have TonB-dependent outer membrane receptors (e.g., ZnuD in Neisseria meningitidis), that are regulated by Zur and involved in zinc uptake [22]. It remains unclear in what form zinc is transported via these systems, but it is tempting to postulate that they may deliver “zincophores” into the cell. More recently, a type VI secretion system in both Burkholderia thailandenis and Yersinia pseudotuberculosis was found to play a role in scavenging zinc specifically during oxidative stress [23, 24].
Here we report the effect of zinc-depletion on growth, cellular zinc quotas, and gene transcription in B. cenocepacia H111.
Materials and Methods
Bacterial Strains and Growth Conditions
Burkholderia cenocepacia H111 was obtained from the Belgian co-ordinated collection of microorganisms. Cultures were grown in either LB-broth (Millers), or a modified Glycerol-glycerophosphate medium (GMM) with or without the addition of zinc [25]. Cultures were routinely grown aerobically at 37°C within an orbital shaking incubator.
The composition of GGM medium was as follows: 40 mM MES, 18.7 mM NH4Cl, 13.4 mM KCl, 7.64 mM β-glycerophosphate, 5.0 mM glycerol, 4.99 mM K2SO4, 1.0 mM MgCl2, 0.134 mM EDTA, 68.0 µM CaCl2·2H20, 18.5 µM FeCl3·6H2O, 12.28 µM ZnSO4·7H2O, 1.62 µM H3BO3, 0.5 µM MnCl2·4H2O, 587 nM CuCl2·2H2O, 344 nM Co(NO3)2·6H2O, and 80.9 nM (NH4)6Mo7O24·4H2O, in MilliQ (18 MΩ cm−1) water. To prepare the medium, bulk components (MES, NH4Cl, KCl, K2SO4, glycerol, H3BO3), dissolved in MilliQ water were passed through a Chelex 100 column (Bio Rad, United Kingdom) to remove contaminating metal cations, following the manufacturer’s instructions. CaCl2 was subsequently added, and the solution sterilised in polycarbonate bottles using a microwave, to avoid introduction of contaminating zinc [26]. After sterilisation, trace metals dissolved in EDTA, MgCl2, and β-glycerophosphate were added by filter sterilisation using 0.2 µm PTFE syringe filters. Syringes without rubber seals were used to minimise zinc contamination. Chemicals were purchased from either Merck or Fisher Scientific and were trace metal grade where available. To further minimise the potential for zinc contamination, culture flasks, measuring cylinders, and other plasticware was washed in 3% trace metal grade nitric acid overnight, and extensively rinsed using MilliQ water (18 MΩ cm−1) before use.
Growth Curves
Ten mL of overnight cultures of B. cenocepacia H111 in LB, were harvested by centrifugation at 3,000 g for 15 min, and the cell pellets washed by gently resuspending in GGM without trace metal additions but supplemented with 1 mM EDTA to remove surface adsorbed metals. Cells were pelleted again and further washed in trace metal free GGM to remove EDTA. After a final centrifugation step, cells were resuspended in 10 ml of complete GGM with or without the addition of zinc. Twenty µL of the resuspended culture was added to 180 µl of fresh GGM in 96-well plates. Plates were sealed using Breathe-Easy® plate seals and incubated at 37°C with orbital shaking at 200 rpm in a CLARIOstar plate reader (BMG LABTECH). Optical density (600 nm) measurements were taken every 10 min for 20 h.
Inductively Coupled Plasma Mass Spectrometry (ICP-MS)
B. cenocepacia H111 was cultured in zinc depleted and zinc replete GGM and harvested at late exponential growth phase, by centrifugation at 3,000 g. Cell pellets were washed with EDTA as described above to remove any surface adsorbed metals. After washing, the cells were collected by centrifugation and dried to constant weight before being digested in 70% trace metal grade nitric acid for 7 days at room temperature. Following complete digestion, samples were diluted to 2% nitric acid in MilliQ water and passed through 0.45 µm PTFE filters. ICP-MS analysis was performed using a NexION 300X instrument (PerkinElmer). Calibration was performed with an external 66Zn standard across the 0–1,000 ppb range. 45Sc was used as the internal standard. The instrument was operated in Helium KED mode to remove any polyatomic interferences. Five technical replicates were taken for each sample analysed, and four biological replicates were performed.
RNA-Sequencing
RNA-sequencing and differential gene expression analysis was performed by GENEWIZ™. B. cenocepacia H111 was grown in zinc depleted and zinc replete GGM to mid-exponential growth phase, as described above. Cells were harvested by centrifugation at 3,000 g, and cell pellets stored at −80°C for 1 week before shipping on dry-ice to GENEWIZ™ for RNA extraction, RNA library preparation and RNA-sequencing. Total RNA was extracted using a Qiagen RNeasy Plus Mini Kit. rRNA depletion was performed using Qiagen FastSelect rRNA bacteria kit. RNA quantity and integrity were determined using a Qubit 2.0 Fluorometer, and Agilent 4200 Tapestation. RNA sequencing libraries of 150 bp reads, were prepared using a NEBNext Ultra II RNA library kit. Samples were paired end sequenced using an Illumina Hiseq instrument.
Data Processing and Differential Gene Expression Analysis
Data processing was performed by GENEWIZ™. RNA-sequence reads were trimmed using Trimmomatic v. 0.36, removing adapter sequences. The trimmed reads were mapped to the B. cenocepacia H111 reference genome [27], using STAR aligner v. 2.5.2b. Unique gene hit counts were calculated by using featureCounts v.1.5.2. Only unique reads within exon regions were counted. Gene hit counts were used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between cells grown in zinc replete and zinc depleted GGM was performed. p-values and Log2 fold changes were calculated using the Wald test. Genes with adjusted p-values < 0.05 and absolute log2 fold changes > 1 were called as being differentially expressed.
RT-qPCR
RNA was extracted from cell pellets using a GeneJET RNA Purification kit (Thermo Scientific™) and used a template for RT-qPCR. Gene sequences were extracted from the Burkholderia genome database [27], and primers designed using Primer3 v. 4.1.0 [28]. A full list of primers used is given in Supplementary Table S1. RT-qPCR was performed using Power SYBR™ green RNA-to-CT 1-step kit (Applied Biosytems™), following the manufacturer’s instructions. PCR was performed using a Quantstudio 3 real time PCR system (Applied Biosystems™). Cycling was as follows: 1 cycle of 48°C for 30 min, 1 cycle of 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 60 s. Relative levels of gene expression were calculated using the 2−ΔΔCt method using the gyrB gene as an internal control.
Results
Effect of Zinc Depletion on Growth of B. cenocepacia H111
B. cenocepacia H111 was cultured in zinc replete and depleted GGM and growth monitored using OD600 measurements as a proxy for cell numbers. The data (Figure 1A) reveals no significant difference in growth between the two conditions evaluated. The duration of the lag phase is comparable, and cells grown in zinc depleted and replete media reached similar final cell numbers with an average OD600 of 0.54 and 0.59, respectively at 20 h. Average doubling times during exponential growth across both conditions was 226 min. That compares to a doubling time in LB-broth of 108 min (data not shown). Growth rates in LB in our lab are comparable to previously reported doubling times for Bcc species of between 70–186 min [29]. The result of this experiment is significant for two reasons. Firstly, the data shows that B. cenocepacia H111 is well adapted for growth in zinc-depleted environments such as those expected to be encountered in the host, and secondly because it demonstrates that GGM-medium is suitable for trace metal studies in Burkholderia species more generally.
FIGURE 1
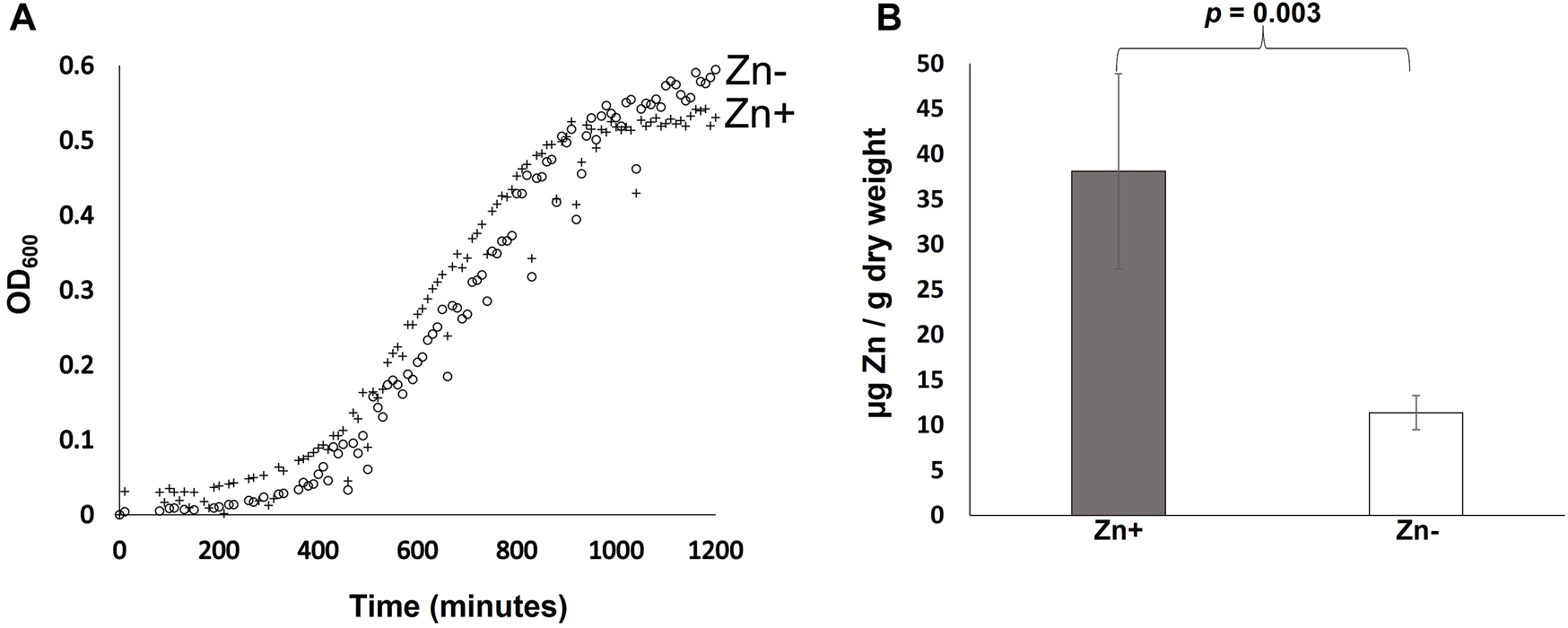
Effect of zinc availability on growth and cellular zinc quotas of B. cenocepacia H111. (A) Growth of B. cenocepacia H111 in zinc-replete GGM (crosses), and zinc-depleted GGM (open circles), over 20 h. OD600 values measured as a proxy for cell numbers. Mean values from 12 replicate samples are plotted. (B). Total cellular zinc quotas (expressed as µg zinc per g of dry cell weight) in B. cenocepacia H111 following culture in zinc-replete GGM (grey bar), and zinc-depleted GGM (white bar). Mean values from 4 independent biological replicates are plotted, with error bars showing standard deviations. The p-value from a two-tailed t-test is given.
Effect of Zinc Depletion on Total Cellular Zinc Quotas
The effect of zinc availability on total cellular zinc quotas was determined using Inductively coupled plasma mass spectrometry (ICP-MS). The data shows that that cellular zinc quotas are significantly reduced when B. cenocepacia H111 cells are grown in zinc depleted GGM compared to when they are grown in the replete medium (Figure 1B). Cells grown in the zinc replete GGM contained an average of 38.10 µg of zinc per g of dry cell weight, compared to just 11.35 µg of zinc per g of dry cell weight in cells grown in the zinc depleted medium. This result is surprising when taken in the context of the previous growth experiment that showed zinc depletion had no measurable effect on growth rates (Figure 1A), and together, these data suggest that B. cenocepacia H111 may either maintain significant zinc stores within the cell during replete conditions, or use “zinc sparing” as a mechanism to reduce overall cellular demand for zinc, when bioavailability is low. This phenomenon has been observed in other bacterial species where the expression of genes encoding non-essential zinc dependent proteins is repressed in response to zinc depletion [30].
Changes in Gene Expression in Response to Zinc Depletion
RNA-sequencing was used to measure any changes in gene expression that occur in B. cenocepacia H111 in response to zinc depletion. A total of 201 genes showed significant differential expression, with 85 upregulated and 116 downregulated genes (Figure 2), representing 2.95% of predicted genes. The full list of significantly differentially expressed genes is given in Supplementary Table S2, and the 33 genes that had more than a Log2 fold increase in expression, are also listed in Table 1. The gene with the greatest fold increase in expression (locus tag I35_RS28540) encodes a hypothetical cytoplasmic protein [27]. A protein BLAST of the translated gene sequence reveals that the gene encodes a protein with significant sequence similarity to members of the COG0523 subfamily of P-loop GTPases that are predicted to play a role in metal homeostasis [31]. In the opportunistic pathogen Acinetobacter baumannii, the orthologous gene zigA is zinc binding and required for growth under zinc limited conditions [32]. ZigA from A. baumannii and the putative COG0523 protein from B. cenocepacia H111 share 75% sequence identity, and a conserved metal binding motif (CICC). Closer examination of the genomic neighbourhood of the COG0523 gene in B. cenocepacia, reveals that it sits in a cluster of several genes that are upregulated in response to zinc depletion (Figure 3; Table 1; Supplementary Table S2), and includes genes that encode the usually zinc dependent proteins carbonic anhydrase, dihydrooratase, and 6-carboxy-5,6,7,8-tetrahydropterin synthase (QueD), as well as FolE2, and Acyl-coA dehydrogenase.This data points to a key role for COG0523 in the response of B. cenocepacia to zinc limitation.
FIGURE 2
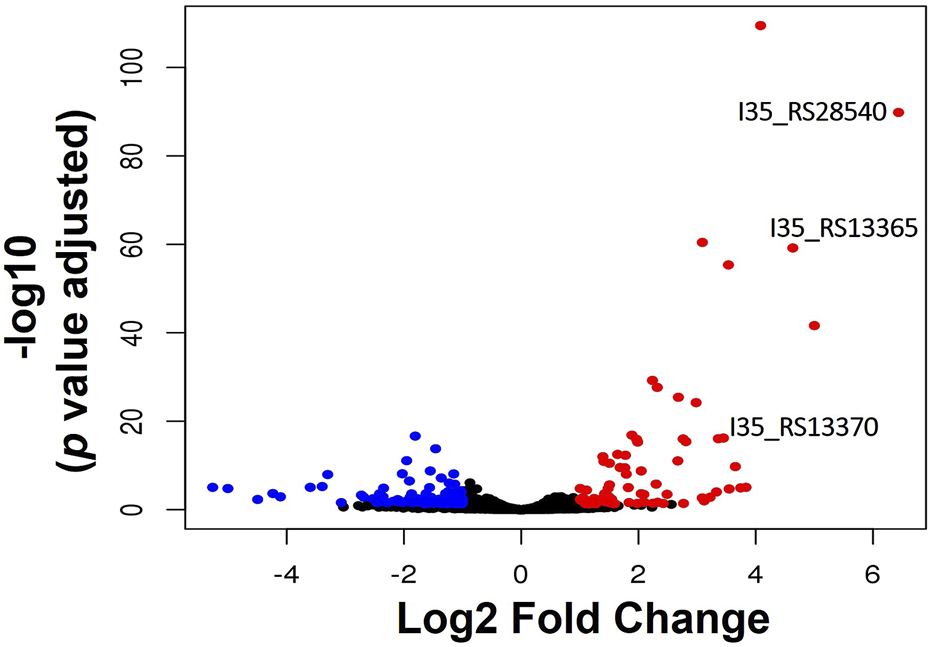
Transcriptional response of B. cenocepacia H111 to zinc depletion. Volcano plot showing the number of genes significantly upregulated (red circles), or downregulated (blue circles) in B. cenocepacia H111 in response to zinc depletion. Genes encoding a putative zinc metallochaperone (locus-tag I35_RS28540), of the COG0523 subfamily of P-loop GTPases, putative Zur-transcriptional regulator (locus-tag I35_RS13365), and putative ZnuA periplasmic Zn-binding protein (locus-tahgI35_RS13370) are indicated.
TABLE 1
| Gene locus | Gene product (from the Burkholderia genome database) [27] | Log2 fold change | p-value adjusted |
|---|---|---|---|
| I35_RS28540 | Hypothetical protein | 6.436363914 | 1.52E−90 |
| I35_RS16050 | Alpha/beta hydrolase | 5.000537573 | 2.42E−42 |
| I35_RS13365 | Fur family transcriptional regulator | 4.633320929 | 6.65E−60 |
| I35_RS28535 | GTP cyclohydrolase | 4.083416486 | 3.42E−110 |
| I35_RS01025 | Phenylacetic acid degradation protein | 3.833347834 | 0.00000913 |
| I35_RS10880 | Histidine ammonia-lyase | 3.743017919 | 0.0000121 |
| I35_RS01000 | 4-hydroxypehnylpyruvate dioxygenase | 3.652890929 | 1.84E−10 |
| I35_RS01030 | Phenylacetate-CoA oxygenase subunit PaaJ | 3.547319093 | 0.0000209 |
| I35_RS28530 | 6-carboxy-5,6,7,8-tetrahydropterin synthase | 3.534715892 | 4.48E−56 |
| I35_RS13370 | Cation ABC transporter substrate-binding protein | 3.449536319 | 6.4E−17 |
| I35_RS16055 | Amino acid ABC transporter ATP-binding protein | 3.362442596 | 8.91E−17 |
| I35_RS10875 | Histidine utilization repressor | 3.330074112 | 0.0000992 |
| I35_RS01035 | Phenylacetic acid degradation protein | 3221187039 | 0.001583599 |
| I35_RS10870 | Urocanate hydratase | 3.121275534 | 0.010056873 |
| I35_RS28525 | Carbonate dehydratase | 3.090073095 | 3.69E−61 |
| I35_RS01040 | Phenylacetate-CoA oxygenase subunit PaaB | 3.088767817 | 0.002296074 |
| I35_RS13375 | ABC transporter ATP-binding protein | 2.983146959 | 6.31E−25 |
| I35_RS28545 | Acyl-CoA dehydrogenase | 2.809792771 | 4.11E−16 |
| I35_RS13620 | MFS transporter | 2.768646664 | 0.039419489 |
| I35_RS06170 | TonB-dependent receptor | 2.762808558 | 1.03E−16 |
| I35_RS29380 | Glyoxalase | 2.680789605 | 3.92E−26 |
| I35_RS23575 | TonB-dependent receptor | 2.48708683 | 0.000346391 |
| I35_RS18920 | Hypothetical protein | 2.423450935 | 0.039764095 |
| I35_RS10860 | Imidazolonepropionase | 2.326828435 | 0.022280782 |
| I35_RS06165 | Phenol degradation protein | 2.320499563 | 2.27E−28 |
| I35_RS22340 | Hypothetical protein | 2.300982216 | 0.00000167 |
| I35_RS20825 | Alpha/beta hydrolase | 2.246852811 | 0.035832439 |
| I35_RS13380 | ABC transporter permease | 2.238628283 | 5.97E−30 |
| I35_RS10865 | Hypothetical protein | 2.099694274 | 0.022469745 |
| I35_RS16595 | DNA-binding protein | 2.091617711 | 0.000362322 |
| I35_RS29435 | Hypothetical protein | 2.072176035 | 0.022280782 |
| I35_RS27035 | 2,2-Dialkylglycine decarboxylase | 2.047152459 | 0.00024177 |
| I35_RS16045 | Hypothetical protein | 2.046747087 | 1.75E−09 |
List of genes with more than Log2-fold increase in expression in response to zinc depletion.
FIGURE 3
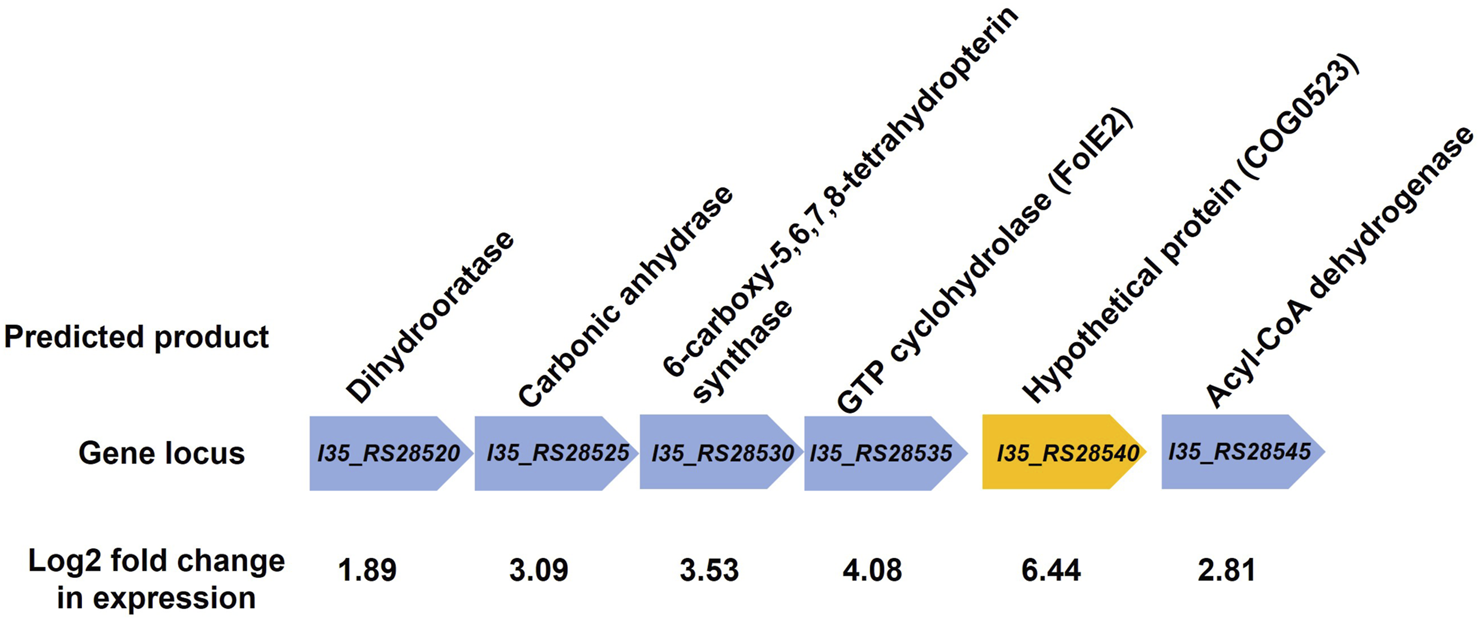
Genomic neighbourhood of putative COG0523 gene. Gene encoding the putative COG0523 zinc metallochaperone is shown (orange arrow), along with neighbouring genes. Gene locus tag is given along with the predicted gene product from the Burkholderia genome database [27]. All genes shown are upregulated in response to zinc depletion, and the Log2 fold change in gene expression is shown.
The RNA-seq data also revealed a set of upregulated genes encoding a “cation ABC transporter,” and a “Fur-family transcriptional regulator”, which based on protein BLAST searches are likely to encode the high affinity zinc uptake transporter ZnuABC and the zinc transcriptional repressor protein Zur. Figure 4A shows the genomic arrangement of these genes, and a putative Zur binding site that was identified upstream of zur that matches exactly to a consensus Zur-box motif for Burkholderia taken from the RegPrecise database (Figure 4B) [33]. It should be noted that it can be difficult to accurately determine the metal specificity of proteins using bioinformatics, and this is the first experimental evidence that this is the ZnuABC system in B. cenocepacia.
FIGURE 4
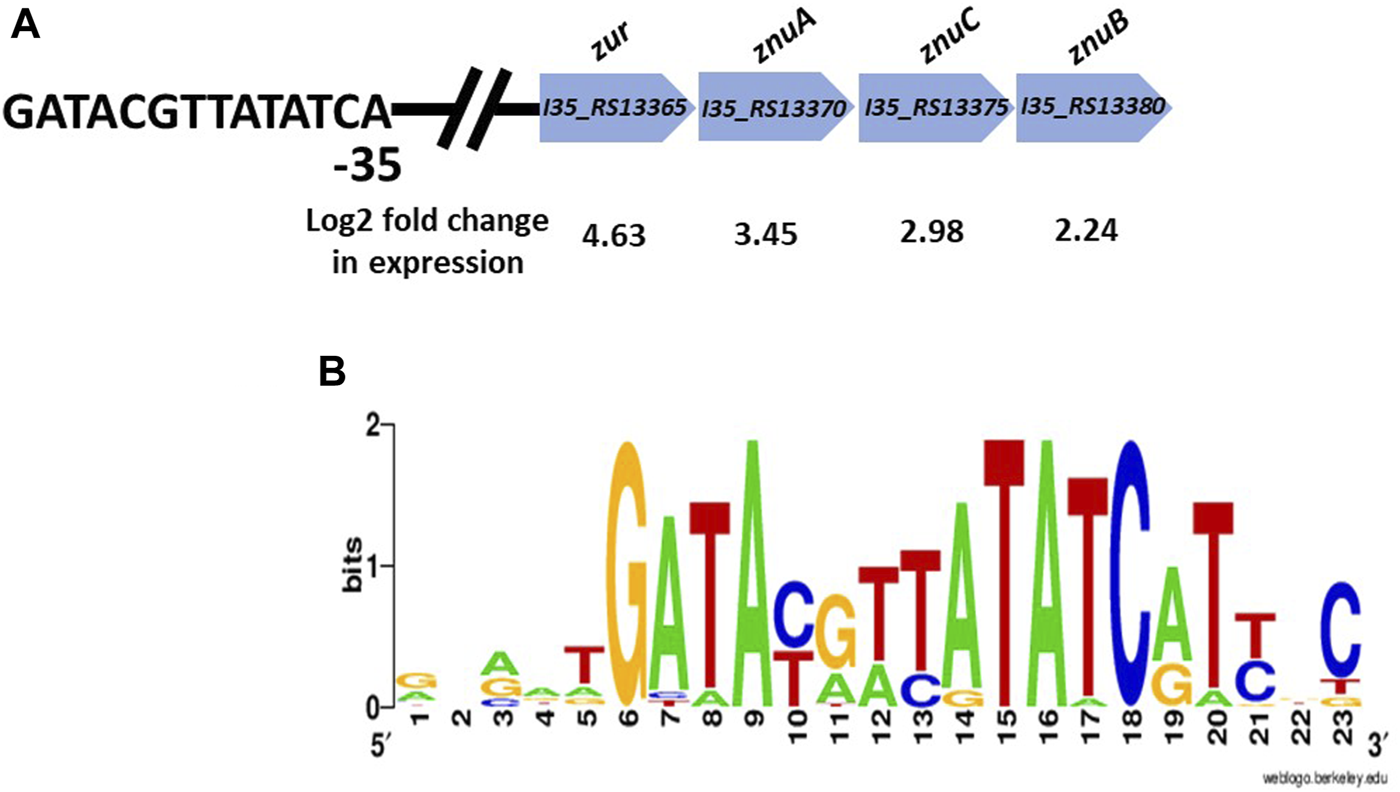
(A) The genomic arragement of the putative znuACB operon in B. cenocepacia H111 is presented, along with the predicted zur gene and Zur-binding site. Log2 fold change in gene expression in response to zinc depletion is shown. (B) Zur-box motif for Burkholderia species taken from the RegPrecise database [33].
Other upregulated genes identified that may play a role in zinc uptake from depleted environments include two TonB-dependent outer membrane receptors and a Major Facilitator Superfamily (MFS) transporter (Table 1), although a specific role for these transporters in zinc uptake remains to be experimentally confirmed.
Intriguingly, the expression of genes related to phenylacetic acid degradation were significantly upregulated too (Table 1), with this catabolic pathway required for the full pathogenicity of B. cenocepacia in a Caenorhabditis elegans infection model [34].
Amongst the genes that are downregulated in response to zinc depletion are some genes encoding usually zinc dependent proteins including aldolase, peptidase M23 and a dihydroorotase (Table 2; Supplementary Table S2), providing further evidence that B. cenocepacia H111 uses a mechanism of zinc sparing to reduce overall cellular demand for zinc during conditions of zinc deprivation. Also noteworthy was the downregulation of a membrane ion permease and a predicted metal transporting ATPase, possibly to reduce zinc efflux from the cell (Table 2; Supplementary Table S2).
TABLE 2
| Gene locus | Gene product (from the Burkholderia genome database) [27] | Log2 fold change | p-value adjusted |
|---|---|---|---|
| I35_RS05820 | Carbamoyl phosphate synthase small subunit | −2.024863193 | 0.023635014 |
| I35_RS04670 | D-amino acid dehydrogenase | −2.027861444 | 7.77E−09 |
| I35_RS00160 | F0F1 STP synthase subunit C | −2.0890612 | 0.007801604 |
| I35_RS03110 | Anthranilate synthase | −2.095343988 | 0.016002175 |
| I35_RS34475 | LysR-family transcriptional regulator | −2.107024951 | 0.005429617 |
| I35_RS00155 | ATP synthase FoF1 subunit A | −2.183650426 | 0.013504754 |
| I35_RS07930 | Peptidase M23 | −2.283848351 | 0.027058355 |
| I35_RS08110 | Uroporphyrin-III C-methyltransferase | −2.348103878 | 0.0000143 |
| I35_RS13745 | MIP family channel protein | −2.34927356 | 0.015213303 |
| I35_RS04075 | Hypothetical protein | −2.355094182 | 0.00130666 |
| I35_RS10765 | Glutamine synthetase | −2.366860655 | 0.01636581 |
| I35_RS00165 | ATP synthase subunit B | −2.411303669 | 0.003024043 |
| I35_RS14515 | Dioxygenase | −2.415249229 | 0.000261085 |
| I35_RS32500 | Spermidine synthase | −2.433711956 | 0.001790561 |
| I35_RS03095 | Thioredoxin | −2.460179293 | 0.027058355 |
| I35_RS12715 | Uroporphyrin-III C-methyltransferase | −2.527603678 | 0.003439743 |
| I35_RS16655 | n-acetyl-gamma-glutamyl-phosphate reductase | −2.686444773 | 0.001583599 |
| I35_RS22845 | Lysine transporter LysE | −2.724901267 | 0.000529129 |
| I35_RS10190 | RNA helicase | −3.068648173 | 0.025920137 |
| I35_RS32515 | Hypothetical protein | −3.300579506 | 1.14E−08 |
| I35_RS32510 | Aldolase | −3.393813782 | 0.00000587 |
| I35_RS32505 | Alkylphosphonate utilization protein | −3.59778736 | 0.00000913 |
| I35_RS17295 | Hypothetical protein | −4.106261473 | 0.001250066 |
| I35_RS13740 | Glycerol kinase | −4.235597783 | 0.000223388 |
| I35_RS23825 | Translation initiation factor IF-1 2 | −4.495477483 | 0.005043043 |
| I35_RS25975 | Diaminopimelate decarboxylase | −5.003385836 | 0.0000168 |
| I35_RS13735 | Glycerol-3-phosphate dehydrogenase | −5.263061988 | 0.00000913 |
List of genes with more than Log2 fold reduction in expression in response to zinc depletion.
The RNA-seq data were validated using RT-qPCR to independently measure changes in the expression of six genes in response to zinc depletion. Several of the genes selected (tonB-dependent receptor, COG0523, zur, znuA and znuC), are putative components of the zinc homeostasis system in B. cenocepacia H111 identified in this study. Good agreement was observed between the two data sets (Figure 5).
FIGURE 5
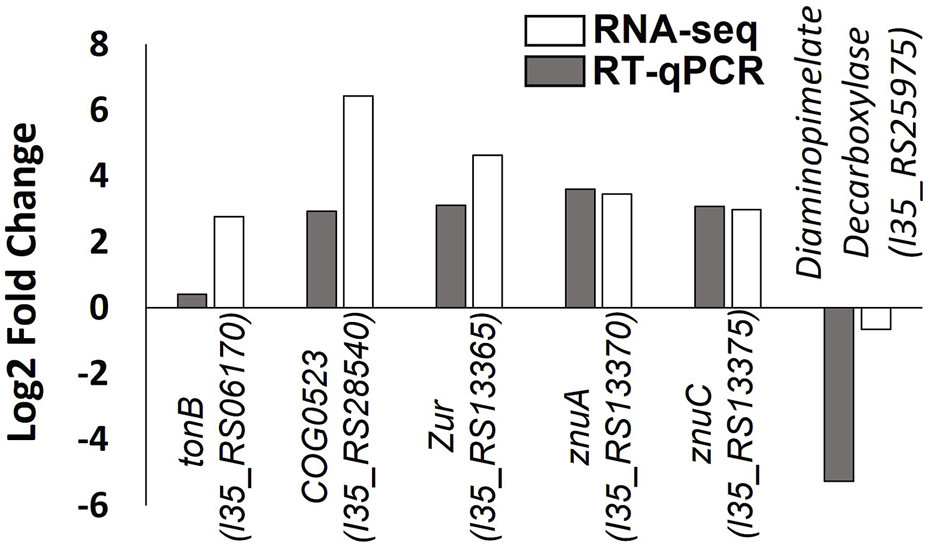
Validation of RNA-seq data by RT-qPCR. Log2 fold change in gene expression of six genes is plotted, with RNA-seq data shown with white bars and RT-qPCR data shown in grey bars. Gene locus tags and protein products are given.
Discussion
Species of bacteria belonging to the B. cepacia complex (Bcc), are most well-known for causing severe respiratory infections in patients with cystic fibrosis and are associated with poor patient outcomes [1–4]. These bacteria have significant intrinsic resistance to major classes of antibiotics, making infections particularly difficult to clear [5, 6]. Despite this and owing to the relatively small numbers of people affected by these bacteria, they have been historically understudied compared to many other human pathogens.
During an infection, our innate immune systems work to starve bacteria of key nutrients including iron and zinc to restrict their growth in the body [13]. Given that zinc and other metals are essential micronutrients for bacteria, metal uptake and homeostasis systems are attractive targets for novel antimicrobial pharmaceuticals. However, a detailed understanding of bacterial metal homeostasis is required first if we are to exploit that vulnerability to tackle infection. In the case of zinc, we are only just starting to understand how bacteria have evolved to survive in zinc poor environments and studies thus far, have been limited to a small number of bacterial species and model organisms. Here we present the first detailed study of the cellular response of any Bcc species to zinc deprivation.
Cellular zinc quotas in B. cenocepacia H111 are significantly reduced when grown in a zinc depleted medium, despite no measurable effect of zinc depletion on growth rates. This is surprising, given that 5%–6% of all proteins in any predicted bacterial genome are thought to bind Zn [10], and with approximately 7,500 protein coding genes, the expectation was that B. cenocepacia H111 would have a relatively high demand for zinc, given its large genome size for a bacterium. This result suggests that B. cenocepacia either maintains significant cellular stores of zinc in replete environments or uses a mechanism of zinc sparing to prioritise limited bioavailable zinc to essential cell functions. The latter is supported by our transcriptomics data, which shows that the expression of several genes encoding usually zinc-dependent proteins are downregulated in response to zinc depletion.
Interestingly, the gene that shows the greatest fold increase in expression when cells are starved of zinc encodes a hypothetical protein, that we have identified as a putative member of the COG0523 subfamily of P-loop GTPases. This is significant as these proteins have been shown to play a role in the response of other bacterial species to zinc limitation, and potentially function as zinc metallochaperones [31, 32]. In B. cenocepacia H111, this gene is located in a cluster of genes that were also upregulated in response to zinc-depletion, and that encode proteins that are usually zinc-binding. Based on this finding we hypothesise that this COG0523 protein plays a key role in the response of B. cenocepacia H111 to zinc deprivation, and that it functions as a zinc-metallochaperone to deliver zinc to essential proteins.
Also upregulated in response to zinc depletion were a number of genes encoding transport proteins, including an ATP-binding cassette transporter that we predict is the ZnuABC Zn2+ importer in this species. The znuABC genes are positioned downstream of a fur-family transcriptional regular, that is the likely zinc-dependent regulator zur, and a regulatory Zur-binding site. It is important to note that it can be difficult to accurately predict the metal specificity of ABC-type transporters from primary sequence data alone, and this is the first experimental evidence that this is the bona fide ZnuABC system in B. cenocepacia H111. Two genes encoding TonB-dependent outer membrane receptors were also significantly upregulated in response to zinc-depletion, and we hypothesise that one or both plays a role in zinc uptake across the outer cell membrane, although further investigation is required to confirm that or otherwise.
Overall, the data reveals that B. cenocepacia responds to zinc depletion in three significant ways, 1) by increasing zinc uptake through the upregulation of genes encoding zinc importers including ZnuABC, 2) by reducing the expression of some non-essential zinc-dependent genes (so called zinc sparing), and 3) through increased expression of a putative zinc metallochaperone of the COG0523 subfamily of P-loop GTPases, presumably to prioritise zinc supply to essential cell functions.
A remaining question concerns effect of zinc bioavailability on the overall virulence and pathogenicity of Burkholderia species, and the potential of pharmaceutical based zinc restriction to tackle Bcc infections. A proof-of-concept study has demonstrated the potential of the periplasmic zinc binding protein ZnuA as a novel drug target in Gram-negative bacteria [35]. In S. enterica serovar Typhimurium, and other Gram-negative pathogens that infect the gastrointestinal tract, ZnuABC is the only high-affinity zinc transporter present. Deletion of znuA or znuABC in Salmonella causes a growth defect in zinc depleted media compared to wild-type strains [35, 36], and the lethal dose of ΔznuABC strains is several orders of magnitude higher than wild-type in a murine model [36]. This underscores the potential of zinc uptake systems as novel targets for antimicrobial drugs in Gram-negative bacteria generally. However, in other species of bacteria redundant zinc importers may be present, reducing the effectiveness of targeting a single protein such as ZnuA. A znuA mutant strain of Pseudomonas aeruginosa, another opportunistic pathogen that like B. cenocepacia colonises the lungs of CF patients, grows efficiently in media containing only trace amounts of zinc, which is likely due to the presence additional mechanisms of zinc uptake [37]. This study is therefore important as it provides the first detailed analysis of zinc-homeostasis in a Bcc species, which is a prerequisite for the development of any novel antimicrobials targeting zinc homeostasis in this species.
In addition to targeting zinc uptake pathways, metal chelating compounds could also be used to target essential zinc dependent enzymes within the cell. A study investigating the mechanism of action of dithiolopyyrolones, found that the antimicrobial properties of these compounds stem from their ability to chelate intracellular zinc [38], and the use of metal chelators as antimicrobials is an active area of research [39].
Summary Table
What Is Known About This Subject
• B. cenocepacia is an opportunistic pathogen, responsible for severe respiratory infections in people with cystic fibrosis.
• These bacteria have significant intrinsic antimicrobial resistance, making infections extremely difficult to treat.
• Bacterial zinc uptake and homeostasis systems are attractive targets for the development of new antibiotic drugs.
What This Paper Adds
• B. cenocepacia is remarkably well adapted for growth in zinc poor environments.
• A putative High-affinity ZnuABC zinc uptake system has been identified in this species.
• A putative zinc chaperone (COG0523) has been identified, that is hypothesised to function in zinc trafficking in the cell.
Summary Sentence
This work represents an advance in biomedical science because it provides the first detailed study of zinc homeostasis in Burkholderia cenocepacia.
Statements
Data availability statement
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB62645.
Ethics statement
Ethics approval was granted by the ethics committee of Birmingham City University. B. cenocepacia is a hazard group 2 microorganism, and the work was authorised by Birmingham City University Biological Safety Committee.
Author contributions
JB as the sole author of this paper, completed the experimental work and wrote the manuscript. RNA-sequencing and differential gene expression analysis was carried out by GeneWiz™.
Funding
This work was funded by a research grant from the Institute of Biomedical Science (United Kingdom), and by the Faculty of Health, Education, and Life Sciences at Birmingham City University, through the pilot project fund.
Acknowledgments
The author is grateful to Mr. Christopher Stark at the University of Birmingham for the ICP-MS analysis.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/bjbs.2023.11597/full#supplementary-material
References
1.
Coenye T Vandamme P Govan JRW LiPuma JJ . Taxonomy and Identification of the Burkholderia Cepacia Complex. J Clin Microbiol (2001) 39:3427–36. 10.1128/JCM.39.10.3427-3436.2001
2.
Isles A Maclusky I Corey M Gold R Prober C Fleming P et al Pseudomonas Cepacia Infection in Cystic Fibrosis: An Emerging Problem. J Pediatr (1984) 104:206–10. 10.1016/s0022-3476(84)80993-2
3.
Drevinek P Mahenthiralingam E . Burkholderia Cenocepacia in Cystic Fibrosis Epidemiology and Molecular Mechanisms of Virulence. Clin Microbiol Infect (2010) 16:821–30. 10.1111/j.1469-0691.2010.03237.x
4.
Govan JR Brown AR Jones AM . Evolving Epidemiology of Pseudomonas Aeruginosa and the Burkholderia Cepacia Complex in Cystic Fibrosis Lung Infection. Future Microbiol (2007) 2:153–64. 10.2217/17460913.2.2.153
5.
Rhodes KA Schweizer HP . Antibiotic Resistance in Burkholderia Species. Drug Resist Update (2016) 28:82–90. 10.1016/j.drup.2016.07.003
6.
Horsley A Jones AM Lord R . Antibiotic Treatment for Burkholderia Cepacia Complex in People With Cystic Fibrosis Experiencing a Pulmonary Exacerbation. Cochrane Database Syst Rev (2016) 2016:CD009529. 10.1002/14651858.CD009529.pub3
7.
Thomassen MJ Demko CA Doershuk CF Stern RC Klinger JD . Pseudomonas Cepacia: Decrease in Colonization in Patients With Cystic Fibrosis. Am Rev Respir Dis (1986) 134:669–71. 10.1164/arrd.1986.134.4.669
8.
Marciano BE Spalding C Fitzgerald A Mann D Brown T Osgood S et al Common Severe Infections in Chronic Granulomatous Disease. Clin Infect Dis (2015) 60:1176–83. 10.1093/cid/ciu1154
9.
Zurita J Mejia L Zapata S Trueba G Vargas AC Aguirre S et al Healthcare-Associated Respiratory Tract Infection and Colonization in an Intensive Care Unit Caused by Burkholderia Cepacia Isolated in Mouthwash. Int J Infect Dis (2014) 29:96–9. 10.1016/j.ijid.2014.07.016
10.
Andreini C Banci L Bertini I Rosato A . Zinc Through the Three Domains of Life. J Proteome Res (2006) 5:3173–8. 10.1021/pr0603699
11.
Bush K Jacoby GA . Updated Functional Classification of Beta-Lactamases. Antimicrob Agents Chemother (2010) 54:969–76. 10.1128/AAC.01009-09
12.
Mikhaylina A Ksibe AZ Scanlan DJ Blindauer CA . Bacterial Zinc Uptake Regulator Proteins and Their Regulons. Biochem Soc Trans (2018) 46:983–1001. 10.1042/BST20170228
13.
Hood MI Skaar EP . Nutritional Immunity: Transition Metals at the Pathogen-Host Interface. Nat Rev Microbiol (2012) 10:525–37. 10.1038/nrmicro2836
14.
Patzer SI Hantke K . The ZnuABC High-Affinity Zinc Uptake System and Its Regulator Zur in Escherichia Coli. Mol Microbiol (1998) 28:1199–210. 10.1046/j.1365-2958.1998.00883.x
15.
Graham AI Hunt S Stokes SL Bramall N Bunch J Cox AG et al Severe Zinc Depletion of Escherichia Coli: Roles for High Affinity Zinc Binding by ZinT, Zinc Transport and Zinc-Independent Proteins. J Biol Chem (2009) 284:18377–89. 10.1074/jbc.M109.001503
16.
Rosadini CV Gawronski JD Raimunda D Arguello JM Akerley BJ . A Novel Zinc Binding System, ZevAB, Is Critical for Survival of Nontypeable Haemophilus Influenzae in a Murine Lung Infection Model. Infect Immun (2011) 79:3366–76. 10.1128/IAI.05135-11
17.
Grass G Wong MD Rosen BP Smith RL Rensing C . ZupT Is a Zn(II) Uptake System in Escherichia Coli. J Bacteriol (2002) 184:864–6. 10.1128/jb.184.3.864-866.2002
18.
Ghssein G Brutesco C Ouerdane L Fojcik C Izaute A Wang S et al Biosynthesis of a Broad-Spectrum Nicotianamine-Like Metallophore in Staphylococcus Aureus. Science (2016) 352:1105–9. 10.1126/science.aaf1018
19.
Grim KP Francisco BS Radin JN Brazel EB Kelliher JL Solórzano PKP et al The Metallophore Staphylopine Enables Staphylococcus Aureus to Compete With the Host for Zinc and Overcome Nutritional Immunity. mBio (2017) 8:e01281–17. 10.1128/mBio.01281-17
20.
Bobrov AG Kirillina O Fetherston JD Miller MC Burlison JA Perry RD . The Yersinia Pestis Siderophore, Yersiniabactin, and the ZnuABC System Both Contribute to Zinc Acquisition and the Development of Lethal Septicaemic Plague in Mice. Mol Microbiol (2014) 93:759–75. 10.1111/mmi.12693
21.
Lhospice S Gomez NO Ouerdane L Brutesco C Ghssein G Hajjar C et al Pseudomonas Aeruginosa Zinc Uptake in Chelating Environment Is Primarily Mediated by the Metallophore Pseudopaline. Sci Rep (2017) 7:17132. 10.1038/s41598-017-16765-9
22.
Stork M Bos MP Jongerius I de Kok N Schilders I Weynants VE et al An Outer Membrane Receptor of Neisseria Meningitidis Involved in Zinc Acquisition With Vaccine Potential. PLoS Pathog (2010) 6:e1000969. 10.1371/journal.ppat.1000969
23.
Si M Wang Y Zhang B Zhao C Kang Y Bai H et al The Type VI Secretion System Engages a Redox-Regulated Dual-Functional Heme Transporter for Zinc Acquisition. Cell Rep (2017) 20:949–59. 10.1016/j.celrep.2017.06.081
24.
Wang T Si M Song Y Zhu W Gao F Wang Y et al Type VI Secretion System Transports Zn2+ to Combat Multiple Stresses and Host Immunity. PLoS Pathog (2015) 11:e1005020. 10.1371/journal.ppat.1005020
25.
Graham AI Hunt S Stokes SL Bramall N Bunch J Cox AG et al Severe Zinc Depletion of Escherichia Coli. J Biol Chem (2009) 284:18377–89. 10.1074/jbc.M109.001503
26.
Kothari V Patadia M Trivedi N . Microwave Sterilized Media Supports Better Microbial Growth Than Autoclaved Media. Res Biotechnol (2011) 2:63–72.
27.
Winsor GL Khaira B Van Rossum T Lo R Whiteside MD Brinkman FS . The Burkholderia Genome Database: Facilitating Flexible Queries and Comparative Analyses. Bioinformatics (2008) 24:2803–4. 10.1093/bioinformatics/btn524
28.
Untergasser A Cutcutache I Koressaar T Ye J Faircloth BC Remm M et al Primer3 – New Capabilities and Interfaces. Nucleic Acids Res (2012) 40:e115. 10.1093/nar/gks596
29.
Caraher E Duff C Mullen T McKeon S Murphy P Callaghan M et al Invasion and Biofilm Formation of Burkholderia Dolosa Is Comparable With Burkholderia Cenocepacia and Burkholderia Multivorans. J Cystic Fibrosis (2007) 6:49–56. 10.1016/j.jcf.2006.05.007
30.
Makthl N Do H Wendel BM Olsen RJ Helmann JD Musser JM et al Group A Streptococcus AdcR Regulon Participates in Bacterial Defense Against Host-Mediated Zinc Sequestration and Contributes to Virulence. Infect Immun (2020) 88:e00097–20. 10.1128/IAI.00097-20
31.
Haas CE Rodionov DA Kropat J Malasam D Merchant SS de Crecy-Lagard V . A Subset of the Diverse COG0523 Family of Putative Metal Chaperones Is Linked to Zinc Homeostasis in All Kingdoms of Life. BMC Genomics (2009) 10:470. 10.1186/1471-2164-10-470
32.
Nairn BL Lonergan ZR Wang J Braymer JJ Zhang Y Calcutt MW et al The Response of Acinetobacter Baumannii to Zinc Starvation. Cell Host Microbe (2016) 19:826–36. 10.1016/j.chom.2016.05.007
33.
Novichkov PS Kazakov AE Ravcheev DA Leyn SA Kovaleva GY Sutormin RA et al RegPrecise 3.0 – A Resource for Genome-Scale Exploration of Transcriptional Regulation in Bacteria. BMC Genomics (2013) 14:745. 10.1186/1471-2164-14-745
34.
Law RJ Hamlin JNR Sivro A McCorrister SJ Cardama GA Cardona ST . A Functional Phenylacetic Acid Catabolic Pathway Is Required for Full Pathogenicity of Burkholderia Cenocepacia in the Caenorhabditis Elegans Host Model. J Bacteriol (2008) 190:7209–18. 10.1128/JB.00481-08
35.
Khemthong S Nuonming P Nookabkaewb S Sukchawalit R Mongkolsuk S . The Agrobacterium Tumefaciens Atu3184 Gene, a Member of the COG0523 Family of GTPases, Is Regulated by the Transcriptional Repressor Zur. Microbiol Res (2019) 222:14–24. 10.1016/j.micres.2019.02.008
36.
Battistoni A Ammendola S Chiancone E Ilari A . A Novel Antimicrobial Approach Based on the Inhibition of Zinc Uptake in Salmonella Enterica. Future Med Chem (2017) 9:899–910. 10.4155/fmc-2017-0042
37.
Ammendola S Pasquali P Pistoia C Petrucci P Petrarca P Rotilio G et al High-Affinity Zn2+ Uptake System ZnuABC Is Required for Bacterial Zinc Homeostasis in Intracellular Environments and Contributes to the Virulence of Salmonella Enterica. Infect Immun (2007) 75:5867–76. 10.1128/IAI.00559-07
38.
D’Orazio M Mastopasqua MC Cerasi M Pacello F Consalvo A Chirullo B et al The Capability of Pseudomonas Aeruginosa to Recruit Zinc Under Conditions of Limited Metal Availability Is Affected by Inactivation of the ZnuABC Transporter. Metallomics (2015) 7:1023–35. 10.1039/c5mt00017c
39.
Chan AN Shiver AL Wever WJ Razvi SZ Traxler MF Li B et al Role for Dithiolopyrrolones in Disrupting Bacterial Metal Homeostasis. PLoS One (2011) 6:2717–22. 10.1073/pnas.1612810114
Summary
Keywords
Burkholderia , RNA-seq, zinc homeostasis, nutritional immunity, Zur
Citation
Barnett JP (2023) Transcriptional Response of Burkholderia cenocepacia H111 to Severe Zinc Starvation. Br J Biomed Sci 80:11597. doi: 10.3389/bjbs.2023.11597
Received
19 May 2023
Accepted
07 September 2023
Published
26 September 2023
Volume
80 - 2023
Updates
Copyright
© 2023 Barnett.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James Paul Barnett, james.barnett@bcu.ac.uk, orcid.org/0000-0003-1827-036X
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.