Abstract
Purpose: Remdesivir use in COVID-19 is associated with cardiac conduction abnormalities from unclear mechanisms. A proposed mechanism is the bioaccumulation of the intermediate metabolite GS-441524 resulting in exogenous activation of cardiac adenosine A1 due to the structural similarity between adenosine and GS-441524. The prolonged half-life of GS-441524 can result in sustained activation of adenosine A1 receptors. In this study, we used molecular modeling of adenosine, GS-441524 and the adenosine A1 receptor to assess the potential mechanistic association of the proposed mechanism.
Methods: Adenosine and GS-441524 structures were acquired from the PubChem database. Ligand docking was carried out using UCSF Chimera. Models were chosen based on greatest binding affinity and minimum root mean square deviation. Figures of resulting structural models were prepared using UCSF Chimera or PyMOL 2.3.5.
Results: By modeling the interaction between the A1 G protein complex and both adenosine and GS-441524, we found that the proposed mechanism of exogenous A1 receptor activation is feasible based on docking compatibility.
Conclusion: The proposed mechanism of exogenous cardiac A1 receptor activation from bioaccumulation of GS-441524 as a cause of observed cardiac conduction abnormalities with the use of remdesivir in COVID-19 is viable. Further studies are needed to assess causality.
Background
Remdesivir is a nucleoside analog which continues to hold key importance in the management of COVID-19 infection among hospitalized patients (1, 2). Recent literature has reported cardiac conduction abnormalities including PR interval prolongation, QTc prolongation, and profound sinus bradycardia attributed to remdesivir administration (3–7). These effects have been postulated to be a result of exogenous activation of G-protein coupled adenosine A1 receptors by the intermediate metabolite GS-441524(4) due to its prolonged half-life and structural similarity to adenosine as seen in Figure 1.
FIGURE 1
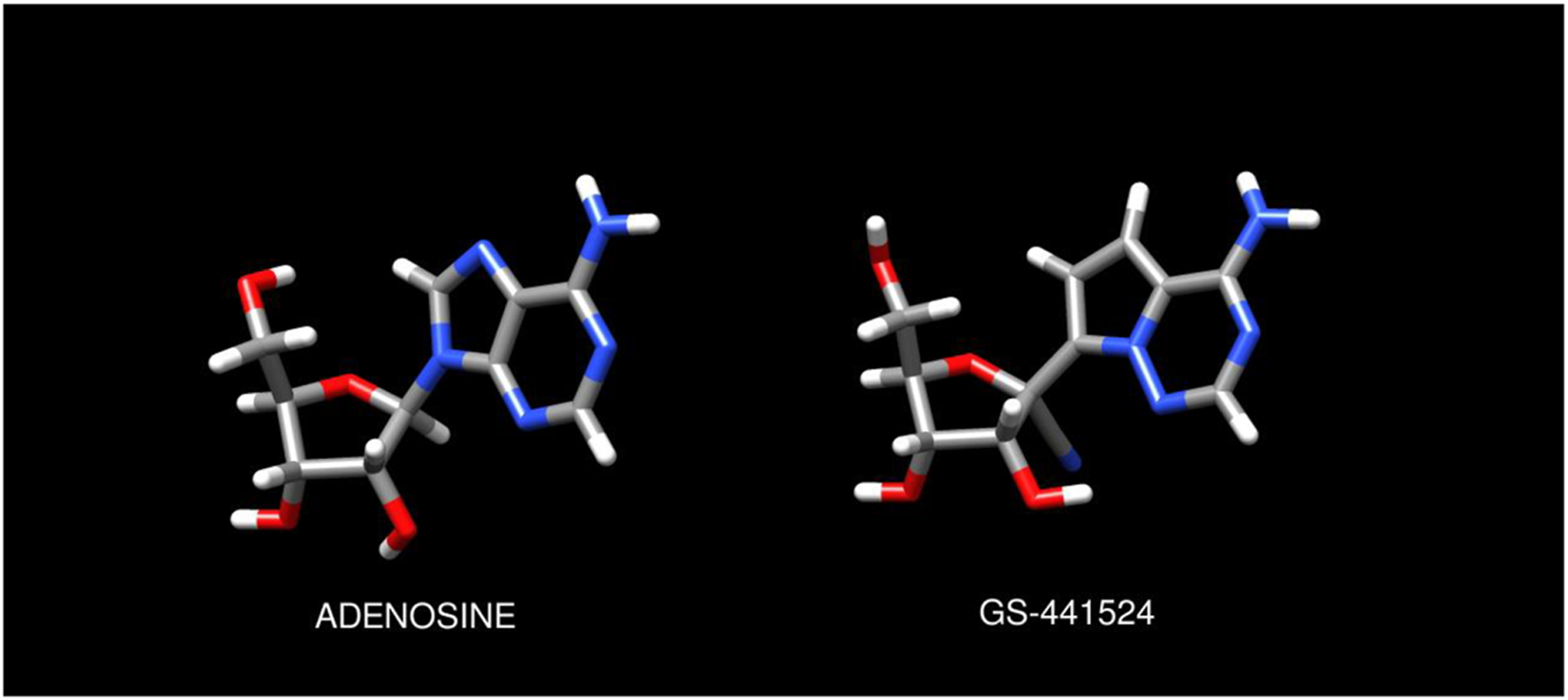
Chemical structures of adenosine and GS-441524 ligands. Molecular models and graphics were created as described in the Methods.
This paper reviews the relevant pharmacologic characteristics of remdesivir, GS-441524, and adenosine and demonstrates that the proposed mechanism is feasible based on molecular modeling of GS-441524 with the cardiac adenosine receptor.
Remdesivir pharmacology, pharmacokinetics
Remdesivir, previously known as GS-5734, is a phosphoramidate prodrug of GS-441524, a 1′-cyano-substituted adenine nucleoside analogue. As a phosphoramidate prodrug, intravenous remdesivir is rapidly hydrolyzed in the serum by extracellular kinases to a nucleoside monophosphate (GS-441524), which then undergoes intracellular conversion to the antiviral, pharmacologically active nucleoside triphosphate metabolite (GS-443902). Remdesivir and GS-441524 are bioisosteres of monophosphates, meaning that they can more quickly be activated and undergo intracellular conversion to the active metabolite. By competing with endogenous nucleosides, GS-443902 can incorporate into viral RNA via inhibition of RNA-dependent RNA polymerase (8–12).
All remdesivir metabolites confer greater selectivity to RNA-dependent RNA polymerase in comparison to human polymerases (8). The presence of exoribonuclease (ExoN) within virus cells acts to correct RNA chain errors to assist in preventing antiviral activity (13). The activity of remdesivir is only minimally affected by ExoNs, as it can more effectively incorporate into viral RNA than other nucleotide analogs (14, 15). This effective incorporation into RNA allows for improved antiviral activity against single-stranded RNA viruses, including Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory distress syndrome coronavirus (SARS-CoV) and SARS-CoV-2 (9, 11).
Upon intravenous administration, remdesivir concentrations decline quickly (half-life ∼1 h) as extracellular kinases work to actively metabolize it to GS-441524 (half-life ∼27 h (11, 16). Remdesivir is highly protein bound (80%–90%), though GS-441524 is not (<5%) (16). It acts as a substrate to several cytochrome (CYP) P450 enzymes in vitro, including CYP2C8, CYP2D6, and CYP3A4. However, the specifics of these pathways are yet to be quantified but are thought to be minor given the rapid metabolism of the prodrug. GS-441524 is not a substrate of major CYP enzymes, suggesting that GS-441524 does not undergo extensive hepatic metabolism (8, 11, 16, 17). Remdesivir byproducts are predominantly excreted via urine, primarily as GS-441524 (49%), followed by remdesivir (10%) and GS-704277 (2.9%). Excretion via feces is negligible (16).
Pharmacologic activity of adenosine on A1 receptors
Adenosine is an endogenous purine nucleoside. It is composed of an adenine molecule with a ribose sugar moiety and is an essential component of adenosine triphosphate (ATP) and cyclic adenosine monophosphate (cAMP) (18, 19). Adenosine activity in the body is widespread, including activity to reduce blood pressure and heart rate, regulation of the sympathetic nervous system, and induction of vasodilation (18).
Adenosine binds to four receptor subtypes that are found on the surfaces of most cells within the body: A1, A2A, A2B, and A3. A1 receptor activation leads to negative chronotropic and dromotropic effect, A2 receptor activation leads to inotropic effects and vasodilation, and A3 receptors are minimally expressed in the myocardium. The adenosine A1 receptors are Gi protein-coupled receptors on cell surfaces with inhibitory functions when activated, leading to negative chronotropic and dromotropic effects (18, 20, 21). A1 receptors are expressed in the brain, spinal cord, kidney, spleen, and heart and have a strong affinity for adenosine (20).
In the heart, adenosine binds to A1 receptors primarily expressed in the atria, leading to decreased cAMP production, inhibition of protein kinase A and voltage-gated calcium channels, and subsequent opening of ATP-sensitive potassium gated channels. The inhibition of this calcium influx and increased potassium current mediates the inhibition of atrioventricular (AV) node conduction, causing a shortened action potential duration with refractoriness (18, 20, 21).
Methods
Ligand binding docking and structural modeling
The human adenosine A1 receptor-Gi2-protein complex (PDB ID: 6D9H) (22) was used as the target receptor. The 3D structural coordinates for both adenosine (compound CID 60961) and GS-441524,(2R,3R,4S, 5R)-2-(4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl)-3,4-dihydroxy-5-(hydroxymethyl)oxolane-2-carbonitrile (Compound CID 44468216) structures were acquired from the PubChem database at the NIH NCBI (23). GS-441524 was the only metabolite tested due to the lack of structural plausibility for the other metabolites. Prior to modeling, both ligand structures were minimized using WebMO (24). Ligand docking was carried out using UCSF Chimera (25) and AutoDock Vina (26). The AutoDock Vina search volume was centered on the A1 adenosine binding site, as described previously (22). The search volume dimensions were set to encompass both the putative binding site and the adjacent trans-membrane helical protein regions. Maximum binding modes was set at ten, with a maximum energy difference of 2 kcal/mol. Models were chosen on the basis of greatest binding affinity and minimum root mean square deviation. Figures of resulting structural models were prepared using UCSF Chimera or PyMOL 2.3.5. Datasets are available upon request: The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ligand binding docking and structural modeling—Results
Our team simulated the interaction between both adenosine and GS-441524 with the A1 receptor. Figure 2 shows the adenosine molecule (purple) binding within the G protein complex.
FIGURE 2
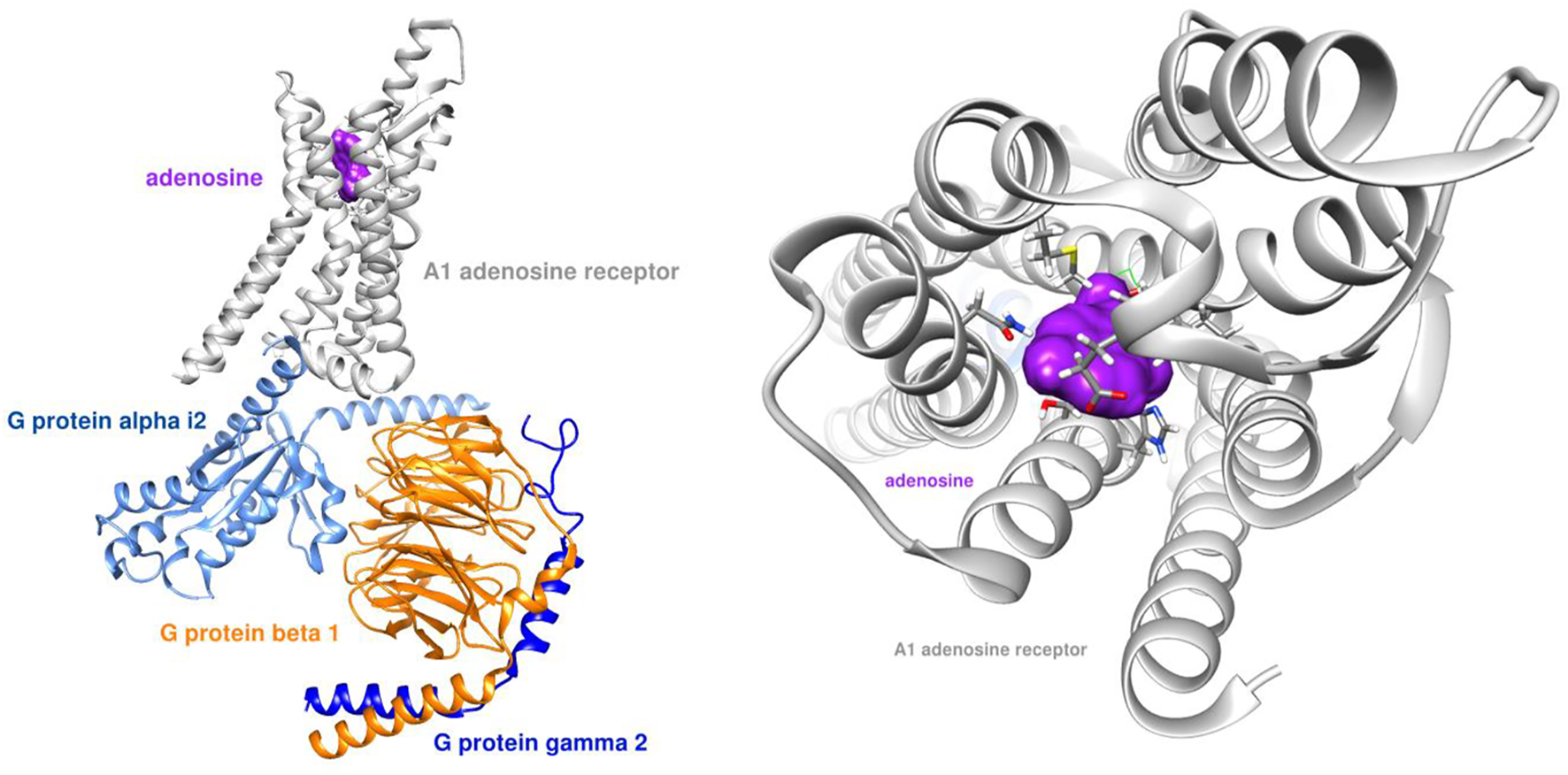
Adenosine receptor - G protein complex (PDB ID: 6D9H) with adenosine ligand.
Figure 3 demonstrates GS-441524 (green) in its pre-docking state with the A1 G protein complex. The adenosine docking site is highlighted in this figure by a green square.
FIGURE 3
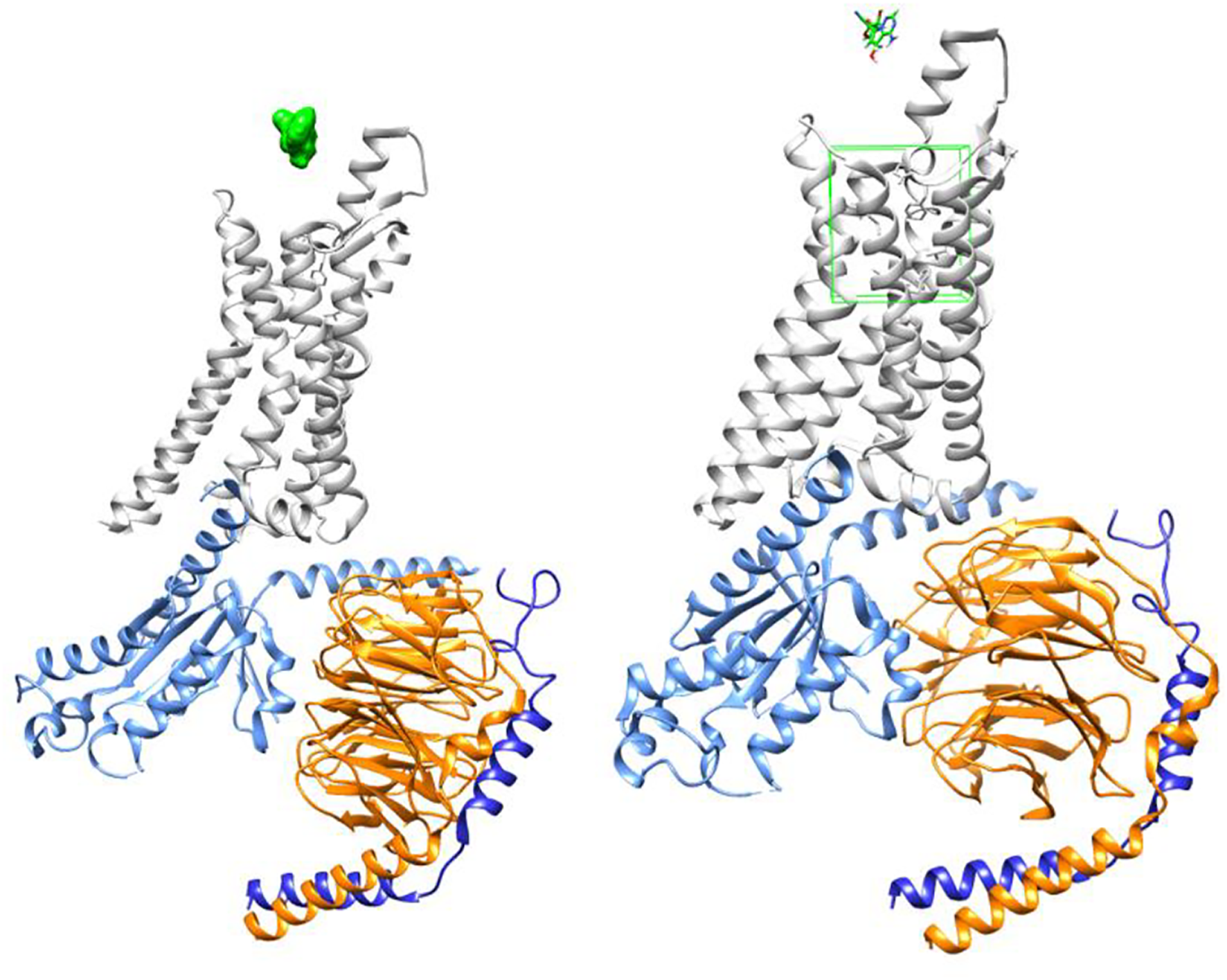
Adenosine receptor - G protein complex (PDB ID: 6D9H) with GS-441524 ligand/Pre-docking.
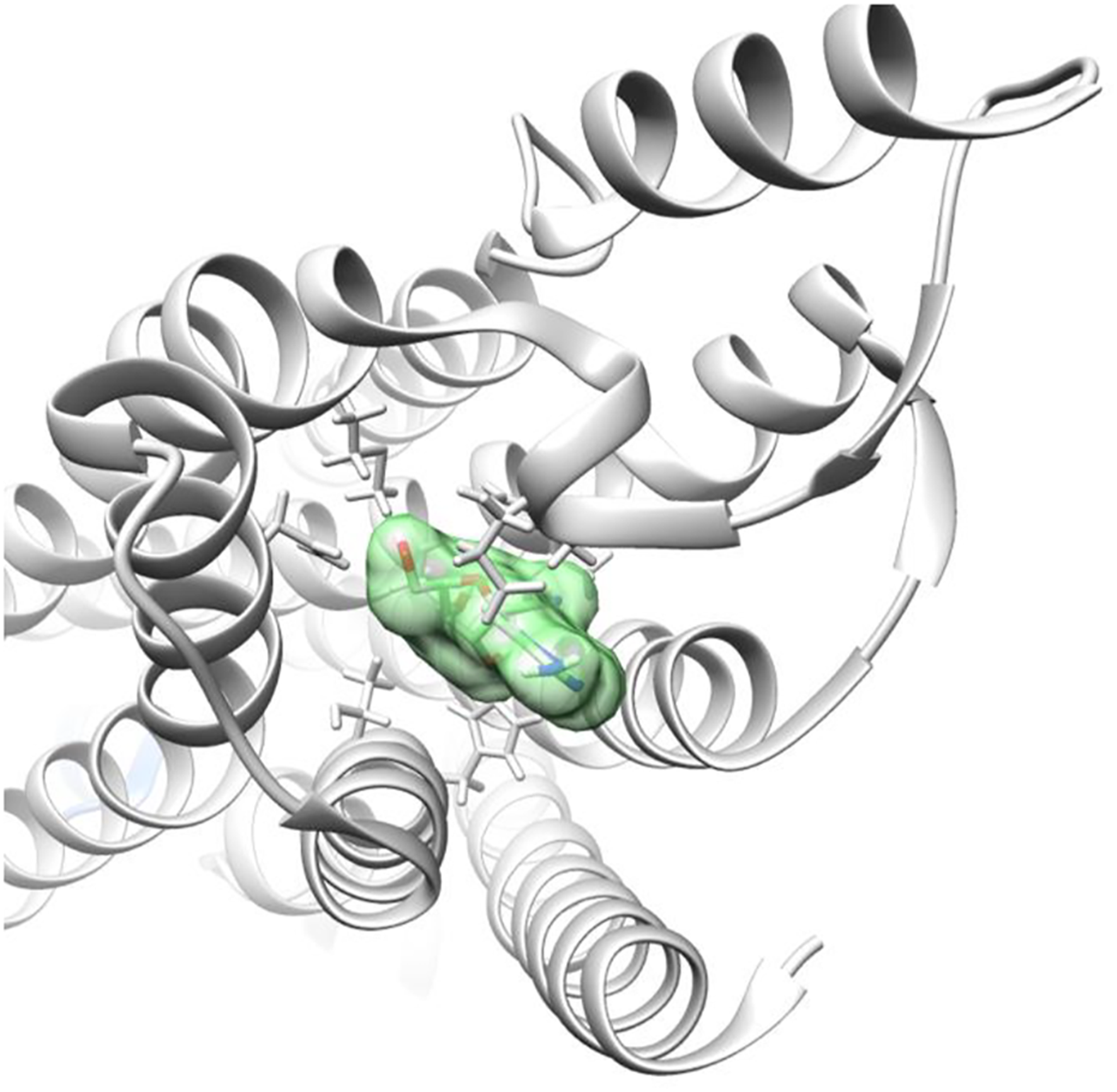
Figures 4, 5 demonstrate GS-441524 (green) in its docking state within the A1 G protein complex.
FIGURE 4
Adenosine receptor - G protein complex with GS-441524 ligand/Post-docking – Axial view.
FIGURE 5
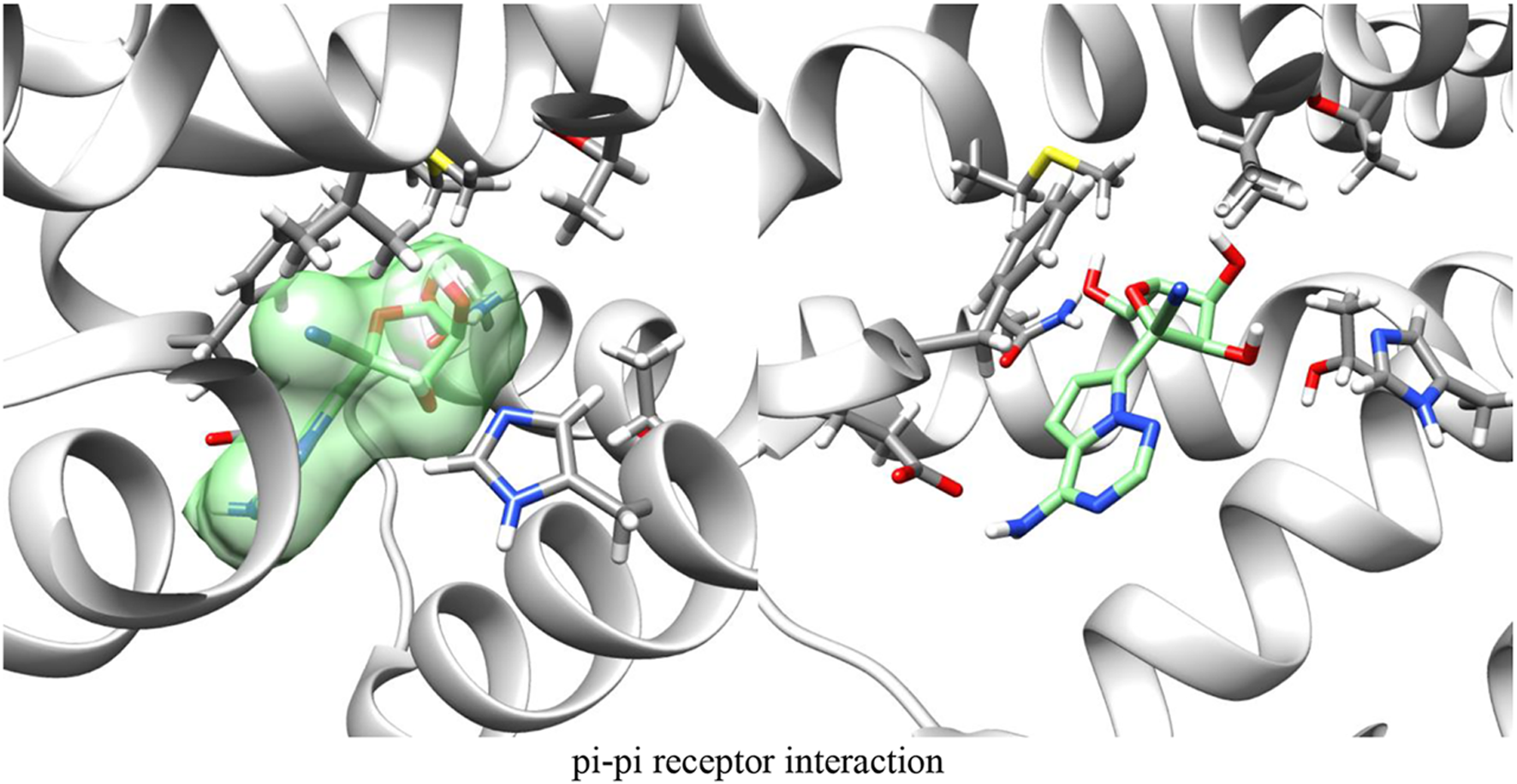
Adenosine receptor - G protein complex with GS-441524 ligand post-docking.
By modeling the interaction between the A1 G protein complex and both adenosine and GS-441524, we found that the proposed mechanism of exogenous A1 receptor activation is feasible based on docking compatibility.
Discussion
Molecular modeling described above demonstrates a high level of binding affinity for GS-441524 as an exogenous ligand of the adenosine A1 receptor. Bioaccumulation of GS-441524 is postulated to result in transient, exogenous activation of cardiac adenosine A1 receptors. Stimulation of these receptors is known to have a myocardial depressant effect by slowing conduction and suppressing cardiac pacemaker function (21). This hypothesis is supported by clinical evidence of negative chronotropy, AV conduction delay, ventricular depolarization delay. In our study, we provide supporting evidence for bioaccumulation of GS-441524 as an important factor in the observed incidence of cardiac conduction abnormalities and proarrhythmic effects in patients treated with remdesivir for COVID-19.
There are several potentially important contributors to cardiac conduction delay in these cases and is likely multi factorial. Conduction abnormalities appear to be more common in those with baseline cardiac conduction system disease (e.g., left bundle branch block, right bundle branch block, etc.) (27). Additionally, both hypoxia and inflammation have been shown to induce increased adenosine metabolism and signaling (28, 29). We hypothesize that the well-characterized cytokine-mediated inflammatory state and the hypoxia induced by COVID-19 increases the endogenous adenosine, which in combination with exogenous GS-441524 bioaccumulation results in transient conduction delay. Our modelling approach provides novel insights into the effects of remdesivir and increases the likelihood of a potential mechanistic explanation of cardiac conduction abnormalities observed with COVID-19 and remdesivir use. Further studies are needed to confirm our findings.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
RyK and RoK developed the study design, formed the team and led manuscript development; AO primarily contributed the remdesivir pharmacology and pharmacokinetics section; CR conducted the molecular modeling and described his methods; RC and DP contributed primarily to the pharmacologic activity of remdesivir on A1 receptors and the discussion. All authors reviewed the final manuscript
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
GaribaldiBTWangKRobinsonMLBetzJCaleb AlexanderGAndersenKMet alReal-world effectiveness of remdesivir in adults hospitalized with coronavirus disease 2019 (COVID-19): A retrospective, multicenter comparative effectiveness study. Clin Infect Dis (2022) 75:e516–e524. 10.1093/cid/ciab1035
2.
GottliebRLVacaCEParedesRMeraJWebbBJPerezGet alEarly remdesivir to prevent progression to severe covid-19 in outpatients. New Engl J Med (2022) 386(4):305–15. 10.1056/nejmoa2116846
3.
BarkasFStylaCPBechlioulisAMilionisHLiberopoulosE. Sinus bradycardia associated with remdesivir treatment in COVID-19: A case report and literature review. J Cardiovasc Dev Dis (2021) 8(2):18. 10.3390/jcdd8020018
4.
GubitosaJCKakarPGerulaCNossaHFinkelDWongKet alMarked sinus bradycardia associated with remdesivir in COVID-19: A case and literature review. JACC: Case Rep (2020) 2(14):2260–4. 10.1016/j.jaccas.2020.08.025
5.
GuptaAKParkerBMPriyadarshiVParkerJ. Cardiac adverse events with remdesivir in COVID-19 infection. Cureus (2020) 12(10):e11132–e. 10.7759/cureus.11132
6.
SelvarajVBavishiCPatelSDapaah-AfriyieK. Complete heart block associated with remdesivir in COVID-19: A case report. Eur Heart J - Case Rep (2021) 5(7):ytab200. 10.1093/ehjcr/ytab200
7.
TouafchiaABagheriHCarriéDDurrieuGSommetAChouchanaLet alSerious bradycardia and remdesivir for coronavirus 2019 (COVID-19): A new safety concerns. Clin Microbiol Infect (2021) 27(5):791.e5–8. 10.1016/j.cmi.2021.02.013
8.
JorgensenSCJKebriaeiRDresserLD. Remdesivir: Review of pharmacology, pre‐clinical data, and emerging clinical experience for COVID‐19. Pharmacother J Hum Pharmacol Drug Ther (2020) 40(7):659–71. 10.1002/phar.2429
9.
MalinJJSuárezIPriesnerVFätkenheuerGRybnikerJ. Remdesivir against COVID-19 and other viral diseases. Clin Microbiol Rev (2020) 34(1):e00162–20. 10.1128/cmr.00162-20
10.
SiegelDHuiHCDoerfflerEClarkeMOChunKZhangLet alDiscovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f] [triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J Med Chem (2017) 60(5):1648–61. 10.1021/acs.jmedchem.6b01594
11.
TempestilliMCaputiPAvataneoVNotariSForiniOScorzoliniLet alCOVID 19 INMI Study Group. Pharmacokinetics of remdesivir and GS-441524 in two critically ill patients who recovered from COVID-19. J Antimicrob Chemother (2020) 75(10):2977–80. 10.1093/jac/dkaa239
12.
YanVCMullerFL. Advantages of the parent nucleoside GS-441524 over remdesivir for covid-19 treatment. ACS Med Chem Lett (2020) 11(7):1361–6. 10.1021/acsmedchemlett.0c00316
13.
TahirM. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J Med Virol (2021) 93(7):4258–64. 10.1002/jmv.27009
14.
GordonCJTchesnokovEPWoolnerEPerryJKFengJYPorterDPet alRemdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem (2020) 295(20):6785–97. 10.1074/jbc.ra120.013679
15.
GordonCJTchesnokovEPFengJYPorterDPGötteM. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem (2020) 295(15):4773–9. 10.1074/jbc.ac120.013056
16.
Veklury [package insert]. Foster City, California: Gilead Sciences (2022).
17.
DebSReevesAA. Simulation of remdesivir pharmacokinetics and its drug interactions. J Pharm Pharm Sci (2021) 24(3):277–91. 10.18433/jpps32011
18.
LaylandJCarrickDLeeMOldroydKBerryC. Adenosine: Physiology, pharmacology, and clinical applications. JACC: Cardiovasc Interventions (2014) 7(6):581–91. 10.1016/j.jcin.2014.02.009
19.
LloydHGEFredholmBB. Involvement of adenosine deaminase and adenosine kinase in regulating extracellular adenosine concentration in rat hippocampal slices. Neurochem Int (1995) 26(4):387–95. 10.1016/0197-0186(94)00144-j
20.
ShryockJCBelardinelliL. Adenosine and adenosine receptors in the cardiovascular system: Biochemistry, physiology, and pharmacology. Am J Cardiol (1997) 79:2–10. 10.1016/s0002-9149(97)00256-7
21.
GuieuRDeharoJ-CMailleBCrottiLTorresaniEBrignoleMet alAdenosine and the cardiovascular system: The good and the bad. J Clin Med (2020) 9(5):1366. 10.3390/jcm9051366
22.
Draper-JoyceCJKhoshoueiMThalDMLiangY-LNguyenATNFurnessSGBet alStructure of the adenosine-bound human adenosine A1 receptor–Gi complex. Nature (2018) 558(7711):559–63. 10.1038/s41586-018-0236-6
23.
KimSChenJChengTGindulyteAHeJHeSet alPubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res (2021) 49(D1):D1388–D1395. 10.1093/nar/gkaa971
24.
PolikWFSchmidtJR. WebMO: Web-based computational chemistry calculations in education and research. WIREs Comput Mol Sci (2022) 12(1):e1554. 10.1002/wcms.1554
25.
PettersenEFGoddardTDHuangCCCouchGSGreenblattDMMengECet alUCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem (2004) 25(13):1605–12. 10.1002/jcc.20084
26.
TrottOOlsonAJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem (2010) 31(2):455–61. 10.1002/jcc.21334
27.
NabatiMParsaeeH. Potential cardiotoxic effects of remdesivir on cardiovascular system: A literature review. Cardiovasc Toxicol (2022) 22(3):268–72. 10.1007/s12012-021-09703-9
28.
BoreaPAGessiSMerighiSVincenziFVaraniK. Pharmacology of adenosine receptors: The state of the art. Physiol Rev (2018) 98(3):1591–625. 10.1152/physrev.00049.2017
29.
BowserJLLeeJWYuanXEltzschigHK. The hypoxia-adenosine link during inflammation. J Appl Physiol (2017) 123(5):1303–20. 10.1152/japplphysiol.00101.2017
Summary
Keywords
remdesivir, conduction abnormalities, COVID-19, adenosine, GS-441524, cardiac conduction abnormalities
Citation
Kingsley R, Rohlman C, Otto A, Chaudhary R, Phelan D and Kirchoff R (2023) Remdesivir-induced conduction abnormalities: A molecular model-based explanation. J. Pharm. Pharm. Sci 26:11208. doi: 10.3389/jpps.2023.11208
Received
19 January 2023
Accepted
31 January 2023
Published
13 February 2023
Volume
26 - 2023
Edited by
Fakhreddin Jamali, University of Alberta, Canada
Updates
Copyright
© 2023 Kingsley, Rohlman, Otto, Chaudhary, Phelan and Kirchoff.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryan Kingsley, kingsley.ryan@mayo.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.