Abstract
Background: Ulcerative colitis (UC) and irritable bowel syndrome (IBS) share various similarities in clinical symptoms, pathogenesis, and treatment. UC concurrent IBS tends toward more severe symptoms and worse prognosis, and promising feasible therapies for the overlapping symptoms remains a challenge. Rhubarb peony decoction (RPD) is a well-known traditional Chinese medicine that has been widely applied in treating UC. RPD may exert extensive therapeutic effects on both IBS and UC. However, the common mechanism of its treatment remains unclear. We aimed to assess the potential pharmacological mechanism of RPD in the treatment of overlapping IBS and UC.
Methods: The active components and targets of RPD were retrieved from ETCM, TCMSP, BATMAN-TCM, and TCM databases. The disease targets were screened by searching the DrugBank, OMIM, TTD, and PharmGKB databases. PPI network analysis was performed and visualized via the STRING platform and Cytoscape software. GO and KEGG enrichment analyses of the hub genes of RPD were predicted to elucidate the potential molecular mechanism. Subsequently, molecular docking was carried out to verify the combination of active compounds with core targets.
Results: By integrating all targets of RPD and disease, a total of 31 bioactive ingredients were identified including quercetin, kaempferol, aloe-emodin, beta-sitosterol, and (+)-catechin, etc. JUN, TP53, MAPK1, RELA, MYC, and ESR1 were explored as potential therapeutic targets among 126 common drug-disease-related targets. They were enriched in the AGE-RAGE signaling pathway in diabetic complications, as well as the NF-kappa B signaling pathway and MAPK signaling pathway. Additionally, some active ingredients were identified as candidates for binding to the hub targets via molecular docking, further suggesting their anti-inflammatory and antioxidative properties.
Conclusion: RPD may exert the overall treatment effect for UC and IBS overlap syndrome via the biological mechanism of “multi-ingredients, multi-targets, and multi-pathways” on inflammation, oxidative stress, immune, oncogenicity, and gut microbiota dysbiosis.
Introduction
Ulcerative colitis (UC) is a chronic idiopathic inflammatory disease that affects the colon, characterized by recurrence and remission of mucosal inflammation. Mucosal inflammation begins in the rectum and continues proximally to the colon [1]. The morbidity of UC is increasing in developing countries, thereby constituting a major public health problem [2]. Irritable Bowel Syndrome (IBS) is one of the most prevalent functional gastrointestinal disorders [3]. It affects approximately 10%∼ 25% of adults worldwide and occurs primarily in women [4, 5]. IBS typically manifests as a chronic and relapsing condition, characterized by several intestinal symptoms including abdominal pain, bloating, a sensation of incomplete evacuation, and altered bowel habits [5, 6]. It has become a healthcare burden on society due to severe psychological state and quality of life (QoL) damage [7]. UC and IBS appear to be quite separate entities, but they do share similarities in clinical symptoms and pathogenesis, even for treatment. Common etiology includes altered gut permeability, gut microbiota dysbiosis, mucosal immune responses, psychological factors, and abnormalities of enteric nerve structure and function [8–10]. Evidence has revealed that around 36% of UC patients have persistent IBS-like symptoms containing abnormalities of sensation, motility, and gut microbiota [11]. Current treatments for IBS include tachykinin, histamine receptor antagonists, and dietary therapy [12]. Therapeutic options for UC include 5-aminosalicylic acid, corticosteroids, anti-TNF agents, and JAK inhibitors [13]. Due to the similarity in pathogenesis, the same treatment may be used for IBS and mild UC patients, including Mesalazine, microecological preparations, and dietary therapy. With the prolongation of western medicine treatment time, distinct degrees of side effects will occur to affect adherence to the treatment process and thus reduce efficacy.
Traditional Chinese medicine (TCM) has been considered an alternative therapy for UC and IBS. TCM is known for its mild nature and has fewer side effects than western medicines. Rhubarb peony decoction (RPD) is a well-known TCM formula, first described in the “Golden Chamber Synopsis” written by the Chinese physician Zhongjing Zhang in the Chinese Eastern Han dynasty, and has more than 2000 years of clinical application history in China. RPD is composed of five herbs: da huang (Rhei Radix et Rhizoma), tao ren (Persicae semen), dong gua zi (Benincasae semen), dan pi (Moutan Cortex), and Mirabilite (a mineral substance, Na2SO4·10H2O). RPD is widely used to treat intestinal carbuncle in China. So far, its novel and effective pharmacological effects on UC have been repeatedly discovered. Luo et al. confirmed that RPD could significantly alleviate the pathological damage in the colon of UC mice by reducing the proportion of Thl7 cells and inflammatory cytokine mediators [14]. Moreover, its major active ingredient, 6-methyl-1,3,8-trihydroxyanthraquinone with the commercial name of Emodin appears to have multiple health benefits in the treatment of a host of diseases, such as immune-inflammatory abnormality, bacterial infections, and tumor progression [15].
Consistent with UC, spleen deficiency and liver stagnation are the root causes of IBS. We speculate that RPD can not only exert significant effects on UC but also IBS. There is a lack of study on the common biological mechanism of RPD in treating IBS and UC. The explanation of the scientific connotation of “the same treatment of different diseases” is one of the significant contents of TCM modernization. Therefore, this study attempted to undertake network pharmacology to further explore the underlying core targets and pathways of the overall treatment effect of RPD against both IBS and UC (Figure 1). Further, this study aimed to provide a reliable reference value for subsequent research including experimental verification and new drug development, as well as for new clinical application.
FIGURE 1
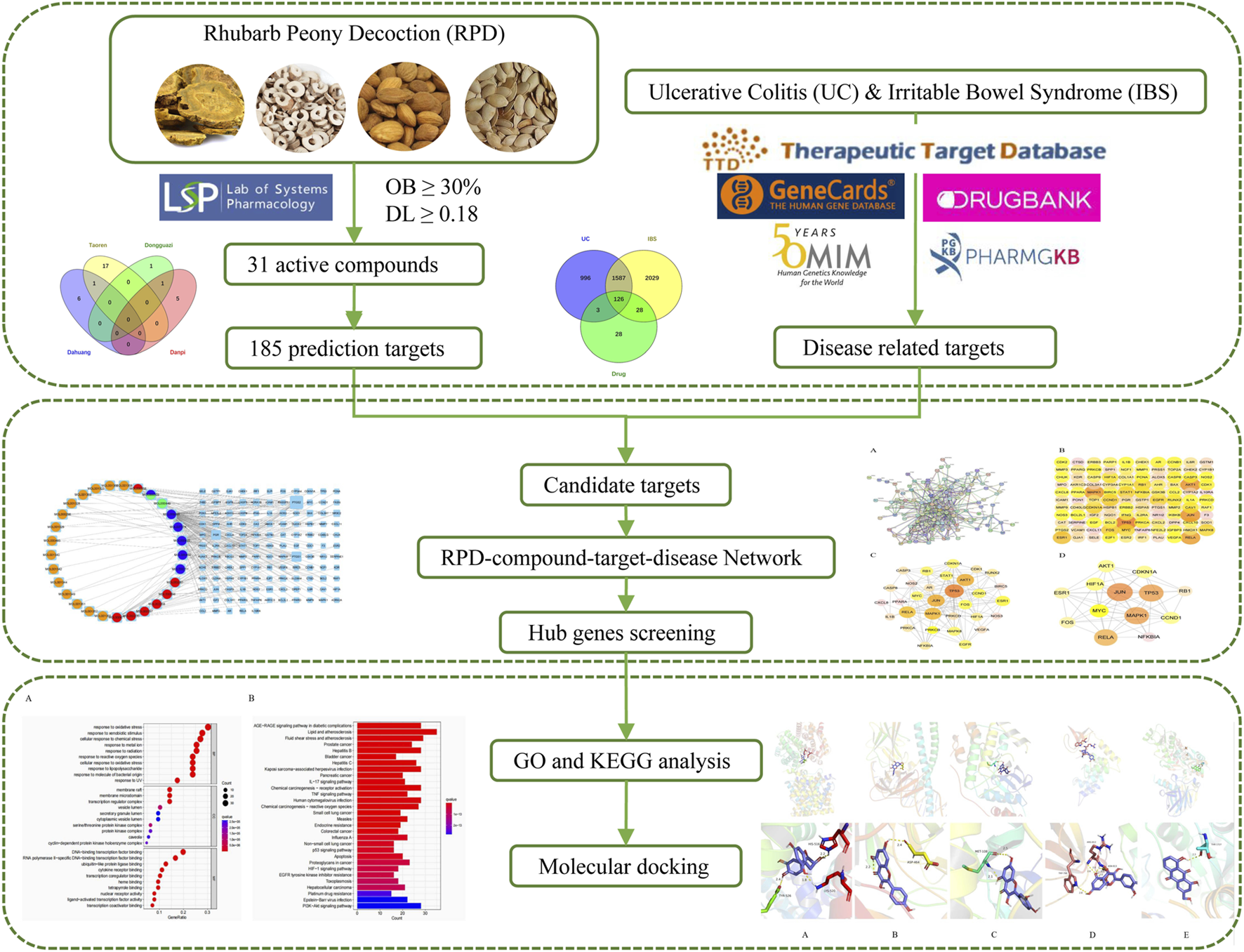
The flowchart of underlining pharmacological mechanisms of RPD against UC and IBS.
Materials and methods
Screening of active components and relative targets in RPD
The active chemical components of five herbs in RPD were retrieved from four public databases including the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform(TCMSP, https://tcmspw.com/tcmsp.php) [16], the Bioinformatics Analysis Tool for Molecular mechanism of Traditional Chinese Medicine (BATMAN-TCM, http://bionet.ncpsb.org.cn/batman-tcm/) [17], the Encyclopedia of Traditional Chinese Medicine (ETCM, http://www.tcmip.cn/ETCM/index.php/Home/Index/index.html) [18], and Traditional Chinese Medicine Database@Taiwan (TCM Database@Taiwan, http://tcm.cmu.edu.tw/) [19]. All of these platforms integrate large-scale authoritative data, including absorption, distribution, metabolism, excretion index scores (ADME parameters), target prediction, and chemical properties. According to the optimal toxicokinetic ADME rules in the TCMSP database, the above TCM compounds were screened as candidate target compounds. Oral bioavailability (OB) ≥ 30% and drug-like (DL) ≥ 0.18 were established as the preset criteria. The collected targets were imported into UniProt (http://www.uniprot.org/) for annotating gene names.
The retrieval of relative disease targets of UC and IBS
In addition, disease-related targets were retrieved from five public databases via the keywords “ulcerative colitis” and “irritable bowel syndrome”: GeneCards (https://www.genecards.org/), Online Mendelian Inheritance in Man (OMIM, https://omim.org/), PharmGKB (https://www.pharmgkb.org/), Therapeutic Target Database (TTD, http://db.idrblab.net/ttd/), and DrugBank (https://go.drugbank.com/). Finally, the common targets of UC and IBS were obtained by merging and eliminating the repeated targets of each disease database.
Constructing the database of intersecting drug-disease target genes
We hypothesized that cross-targeting compounds may be common therapeutic components affecting UC and IBS. Genes that were irrelevant or mismatched between the disease target and RPD were removed. By integrating all targets, the cross-target genes were extracted to construct the assembled target gene library for the following research.
Constructing the protein-protein interaction (PPI) network
The PPI network was constructed and clarified by STRING (https://string-db.org/) [20]. “multiple proteins” was chosen, “Homo sapiens” was set for protein species, and “highest confidence (0.9)” was the minimum interaction threshold. The PPI network was generated to visualize the complex connections. In addition, both active ingredients and corresponding targets were imported into Cytoscape 3.6.1 to further elucidate the interactions of the common target genes in the treatment of UC and IBS with RPD. The CytoNCA plugin was used to calculate the topological parameters of nodes to provide an in-depth analysis of the attributes of nodes in the interactive network. Six main reference parameters were calculated including betweenness centrality (BC), closeness centrality (CC), degree centrality (DC), eigenvector centrality (EC), network centrality (NC), and local average connectivity (LAC). Higher quantitative values of parameters indicate the greater significance of nodes in the network. Target nodes with all six parameters higher than the corresponding median values in the PPI network were selected to construct a new PPI network. The node selection criteria were repeated to obtain the core PPI network.
Enrichment analysis of GO and KEGG
Gene Ontology (GO) term enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were carried out via the R package including “colorspace,” “clusterProfiler,” “dose,” “stringi,” “enrichplot,” and “ggplot2” which were installed in software R 4.2.1. The function “enrichGO” was carried out for GO enrichment analysis, and the database was org. Hs .eg. db (DOI: 10.18129/b9.bioc.org.Hs.eg.db), the “enrichKEGG” function was applied for KEGG enrichment analysis, and the relative database was the KEGG database (https://www.kegg.jp/), respectively. It was determined that the functional terms and pathways with q-values (enrichment score) <0.05 were regarded as statistically significant and retained. The results were sorted according to the q-value, and the top 20 results were visualized as barplot or bubble diagram.
Verification of ingredient–target interaction via molecular docking
The key targets were molecularly docked to the active ingredients for the primary validation. The three-dimensional(3D) structures of key target proteins were downloaded from the Protein Database Bank (https://www.rcsb.org/), and 2D molecular structures of ligands were derived from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), respectively [21]. Chem3D software was utilized to convert ligands into a 3D structure, obtaining the most stable molecular conformation with minimized energy by optimization module. The target protein was presented as a receptor and the water molecule was added with nonpolar hydrogen. The core bioactives and selective proteins were docked by AutoDockVina software and visualized by PyMOL software [22]. The lowest binding energy was regarded as the best-docked conformation, and spontaneous binding occurred while the energy below −5 kJ/mol [23]. A heatmap plot was drawn to visualize these values in R.
Results
Active components and target selection of RPD
Through searching the TCMSP and other online databases to obtain the compounds and prediction targets of RPD. Mirabilite (Na2SO4·10H2O) is a kind of inorganic compound that exerts effects on drug absorption by promoting small intestine movement [24]. After performing ADME screening (OB≥30, DL≥0.18), 31 active ingredients of the main medicine in this prescription were screened (containing 7 types in da huang, 6 types in dan pi, 18 types in tao ren, and 2 types in dong gua zi) (Figure 2A). Using the related target prediction technology in each software to screen the prediction targets of the above-mentioned active ingredients, the repeated targets were eliminated, and finally a total of 185 herbal prediction targets were obtained (Figure 2B) (Table 1, Supplementary Table S1).
FIGURE 2
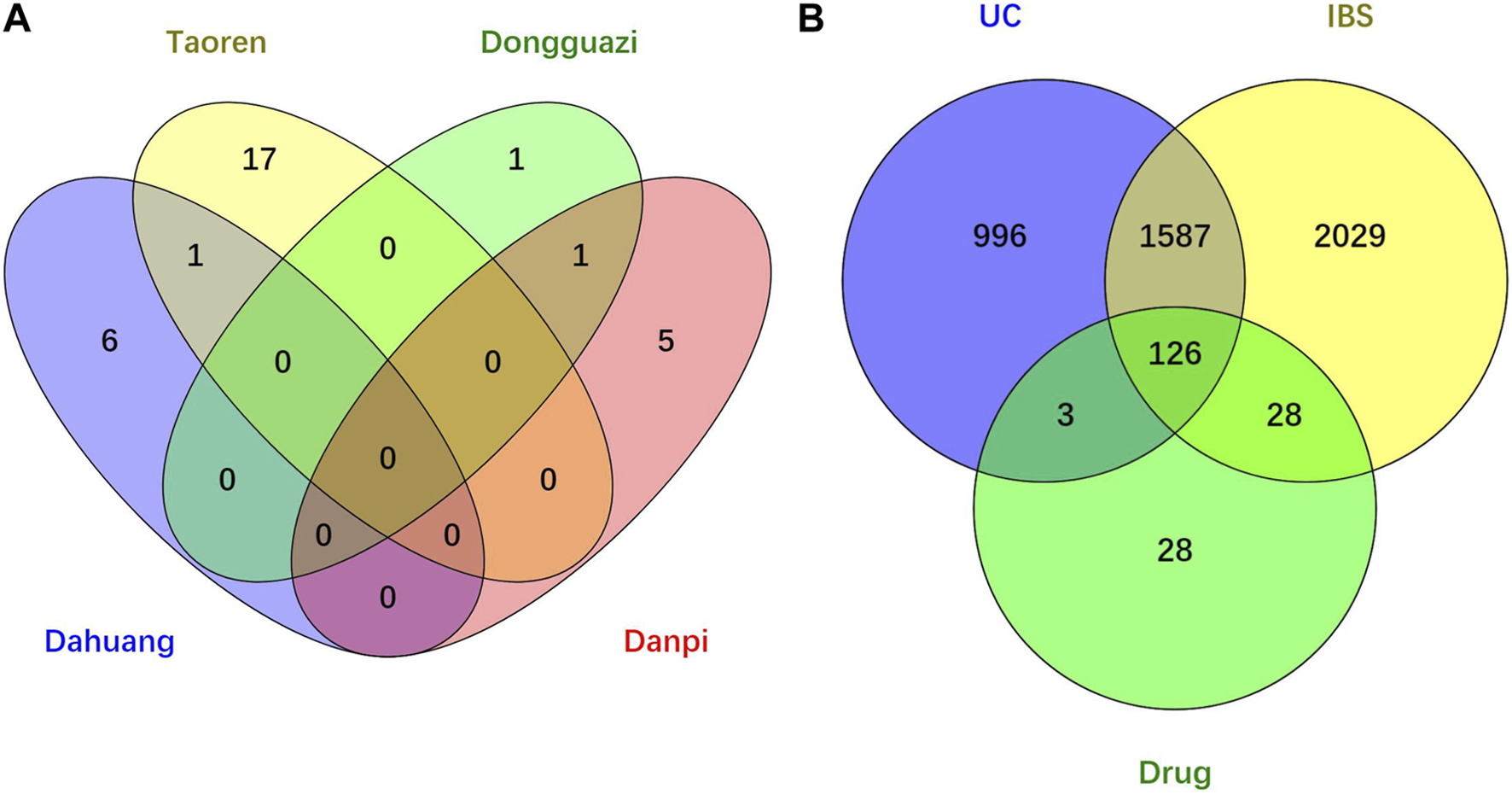
Venn diagram of drug-disease intersection target of RPD for UC and IBS. (A) Core compounds of four herbs from RPD. (B) The common targets of RPD-UC-IBS.
TABLE 1
| Drug | Mol Id | Molecule name | OB (%) | DL |
|---|---|---|---|---|
| dan pi | MOL000211 | Mairin | 55.38 | 0.78 |
| dan pi | MOL000422 | kaempferol | 41.88 | 0.24 |
| dan pi | MOL000492 | (+)-catechin | 54.83 | 0.24 |
| dan pi | MOL007374 | 5-[[5-(4-methoxyphenyl)-2-furyl] methylene] barbituric acid | 43.44 | 0.3 |
| dan pi | MOL000098 | quercetin | 46.43 | 0.28 |
| dan pi/dong gua zi | MOL000359 | sitosterol | 36.91 | 0.75 |
| dong gua zi | MOL000449 | Stigmasterol | 43.83 | 0.76 |
| da huang | MOL002235 | EUPATIN | 50.8 | 0.41 |
| da huang | MOL002268 | Rhein | 47.07 | 0.28 |
| da huang | MOL002281 | Toralactone | 46.46 | 0.24 |
| da huang | MOL002297 | Daucosterol_qt | 35.89 | 0.7 |
| da huang | MOL000358 | beta-sitosterol | 36.91 | 0.75 |
| da huang | MOL000471 | aloe-emodin | 83.38 | 0.24 |
| da huang | MOL000096 | (-)-catechin | 49.68 | 0.24 |
| da huang/tao ren | MOL001358 | gibberellin 7 | 73.8 | 0.5 |
| tao ren | MOL001323 | Sitosterol alpha1 | 43.28 | 0.78 |
| tao ren | MOL001328 | 2,3-didehydro GA70 | 63.29 | 0.5 |
| tao ren | MOL001329 | 2,3-didehydro GA77 | 88.08 | 0.53 |
| tao ren | MOL001340 | GA120 | 84.85 | 0.45 |
| tao ren | MOL001342 | GA121-isolactone | 72.7 | 0.54 |
| tao ren | MOL001344 | GA122-isolactone | 88.11 | 0.54 |
| tao ren | MOL001349 | 4a-formyl-7alpha-hydroxy-1-methyl-8-methylidene-4aalpha,4bbeta-gibbane-1alpha,10beta-dicarboxylic acid | 88.6 | 0.46 |
| tao ren | MOL001351 | Gibberellin A44 | 101.61 | 0.54 |
| tao ren | MOL001352 | GA54 | 64.21 | 0.53 |
| tao ren | MOL001353 | GA60 | 93.17 | 0.53 |
| tao ren | MOL001355 | GA63 | 65.54 | 0.54 |
| tao ren | MOL001360 | GA77 | 87.89 | 0.53 |
| tao ren | MOL001361 | GA87 | 68.85 | 0.57 |
| tao ren | MOL001368 | 3-O-p-coumaroylquinic acid | 37.63 | 0.29 |
| tao ren | MOL000296 | hederagenin | 36.91 | 0.75 |
| tao ren | MOL000493 | campesterol | 37.58 | 0.71 |
Active compounds of RPD.
OB, oral bioavailability; DL, drug-likeness.
Screening disease targets for UC and IBS
We searched the DisGeNET, Drugbank, OMIM, TTD, and PharmGKB databases to obtain the known target genes related to UC and IBS, respectively. Merging each database and deleting the duplicates, we retrieved 3,770 disease targets for IBS and 2,712 disease targets for UC (Supplementary Figures S1, S2). A total of 1713 (63.2% of UC, 45.4% of IBS) disease genes overlap among them, demonstrating they do share similarities in critical pathogenesis. PRD could exert common effects on these two diseases with the explanation of the scientific connotation of “the same treatment of different diseases.” By merging with RPD-related targets, we obtained 154 IBS-related therapeutic targets and 129 UC-related therapeutic targets. Finally, 126 intersecting target genes were considered as the predicted targets for subsequent studies (Table 2). OmicShare online tool was used to construct a Venn diagram for intuitive visual presentation (Figure 2B).
TABLE 2
| No. | Gene | No. | Gene | No. | Gene | No. | Gene |
|---|---|---|---|---|---|---|---|
| 1 | ABCG2 | 33 | COL3A1 | 65 | IFNG | 97 | PLAU |
| 2 | ACHE | 34 | CTSD | 66 | IGF2 | 98 | PON1 |
| 3 | ADH1C | 35 | CXCL10 | 67 | IGFBP3 | 99 | PPARA |
| 4 | ADRA1B | 36 | CXCL11 | 68 | IKBKB | 100 | PPARD |
| 5 | ADRA2A | 37 | CXCL2 | 69 | IL10RA | 101 | PPARG |
| 6 | AHR | 38 | CXCL8 | 70 | IL1A | 102 | PRKCA |
| 7 | AKR1C3 | 39 | CYP1A1 | 71 | IL1B | 103 | PRKCB |
| 8 | AKT1 | 40 | CYP1A2 | 72 | IL2RA | 104 | PRKCD |
| 9 | ALOX5 | 41 | CYP1B1 | 73 | IL6R | 105 | PRSS1 |
| 10 | AR | 42 | CYP3A4 | 74 | IRF1 | 106 | PTGS1 |
| 11 | BAX | 43 | DPEP1 | 75 | JUN | 107 | PTGS2 |
| 12 | BCL2 | 44 | DPP4 | 76 | KDR | 108 | RAF1 |
| 13 | BCL2L1 | 45 | DUOX2 | 77 | MAPK1 | 109 | RASSF1 |
| 14 | BIRC5 | 46 | E2F1 | 78 | MAPK8 | 110 | RB1 |
| 15 | CA2 | 47 | EGF | 79 | MMP1 | 111 | RELA |
| 16 | CASP3 | 48 | EGFR | 80 | MMP2 | 112 | RUNX2 |
| 17 | CASP8 | 49 | ERBB2 | 81 | MMP3 | 113 | SELE |
| 18 | CASP9 | 50 | ERBB3 | 82 | MMP9 | 114 | SERPINE1 |
| 19 | CAT | 51 | ESR1 | 83 | MPO | 115 | SLC6A4 |
| 20 | CAV1 | 52 | ESR2 | 84 | MYC | 116 | SLPI |
| 21 | CCL2 | 53 | F3 | 85 | NCF1 | 117 | SOD1 |
| 22 | CCNB1 | 54 | FASN | 86 | NFE2L2 | 118 | SPP1 |
| 23 | CCND1 | 55 | FOS | 87 | NFKBIA | 119 | STAT1 |
| 24 | CD40LG | 56 | GJA1 | 88 | NOS2 | 120 | THBD |
| 25 | CDK1 | 57 | GSK3B | 89 | NOS3 | 121 | TNFAIP6 |
| 26 | CDK2 | 58 | GSTM1 | 90 | NQO1 | 122 | TOP1 |
| 27 | CDKN1A | 59 | GSTP1 | 91 | NR1I2 | 123 | TOP2A |
| 28 | CHEK1 | 60 | HIF1A | 92 | NR3C2 | 124 | TP53 |
| 29 | CHEK2 | 61 | HMOX1 | 93 | ODC1 | 125 | VCAM1 |
| 30 | CHUK | 62 | HSPA5 | 94 | PARP1 | 126 | VEGFA |
| 31 | CLDN4 | 63 | HSPB1 | 95 | PCNA | ||
| 32 | COL1A1 | 64 | ICAM1 | 96 | PGR |
The intersection targets of RPD and diseases related to UC and IBS.
Pharmacological network construction
The targeting relationship between TCM active ingredients and intersection genes is presented by the TCM compound-disease regulatory network (Figure 3). Based on the “one to multiple,” and “multiple to one” links, the network possessed 155 nodes and 242 edges. The more nodes and edges acquired, the stronger the node interaction. Core genes possessed a higher degree which was positively correlated with node size and edge number. Active ingredients varied in different colors, orange represents tao ren, red represents da huang, blue represents dan pi, and green represents dong gua zi. Quercetin (MOL000098, degree = 103), kaempferol (MOL000422, degree = 37), aloe-emodin (MOL000471, degree = 15), beta-sitosterol (MOL000358, degree = 13), and (+)-catechin (MOL000096, degree = 6) were the top compounds and may be the critical nodes in the network, indicating greater significance anti-UC and IBS effect (Supplementary Table S2). Da huang and dan pi possessed a higher degree value, which is the main medicine in this prescription.
FIGURE 3
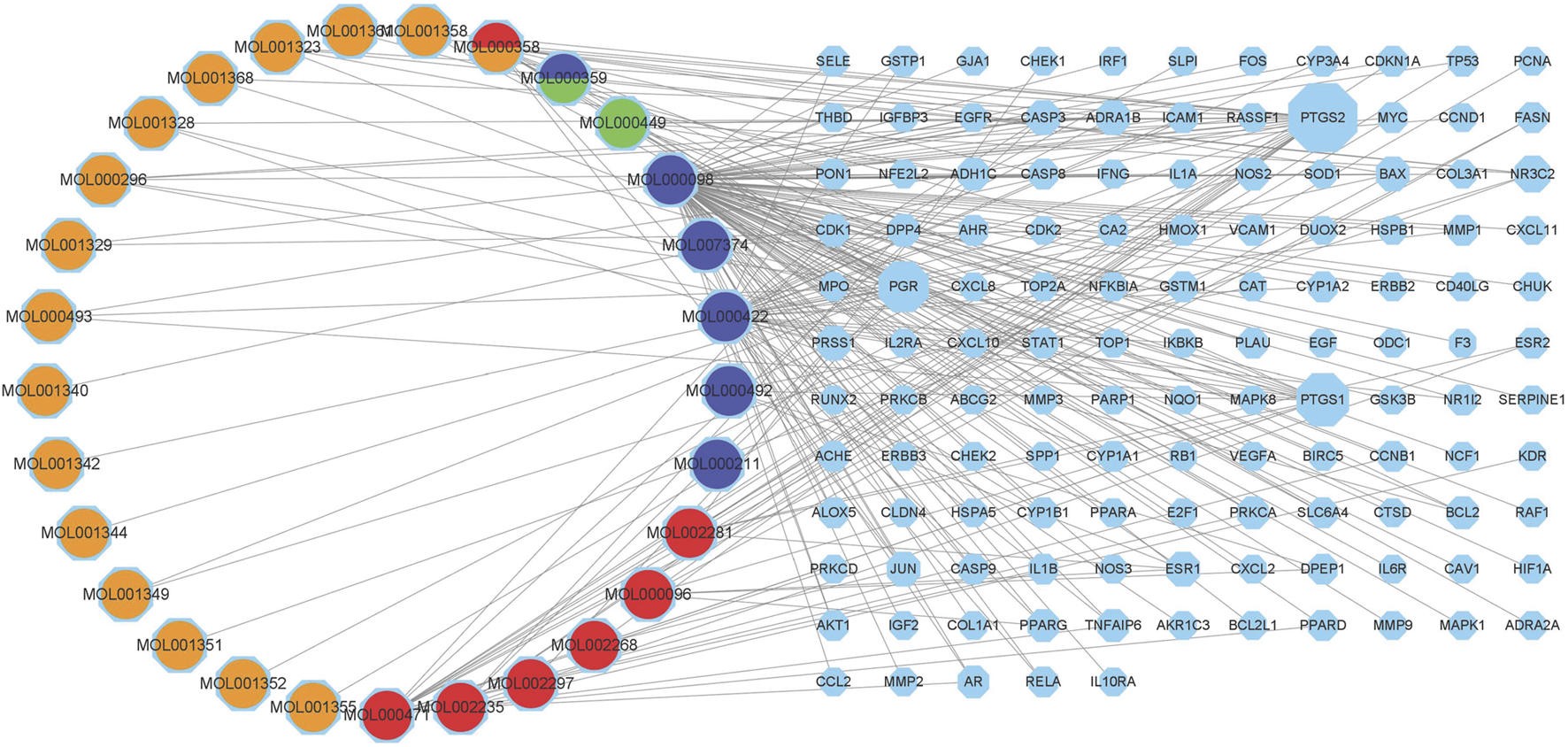
Composition-target network of RPD. This network shows the targeted relationship between the active components of TCM and the intersection of genes. Octagons represent targets. The left Circles stand for compound components. Orange represents tao ren, red represents da huang, blue represents dan pi, and green represents dong gua zi. MOL000358 stands for tao ren and da huang. MOL000359 belongs to dan pi and dong gua zi.
PPI network and topological analysis
The PPI network was constructed to clarify the relationship between RPD and diseases via the STRING software platform, with “the highest confidence (0.9)” for the minimum interaction threshold (Figure 4A). To further visualize and clarify the interactions of proteins, a new PPI network (Figure 4B) comprising 109 nodes and 433 connected edges was performed in Cytoscape 3.8.0. The average node Degree value is 15.89, and the average center clustering coefficient is 0.391. The width of the lines in the interactive PPI network represented the connection between the genes. Subsequently, the CytoNCA plugin was utilized for mining the core goals. The selection criteria were set based on corresponding median values as follows: BC >0, CC >0.4762, DC >6.00, EC >0.0527, NC >4.5714, and LAC >4.00. A new PPI network coinciding with the criteria was extracted with 31 nodes and 153 edges (Figure 4C). Likewise, the core PPI network that met the criteria (BC >13.837, CC >0.6, DC >20.00, EC >0.1645, NC >12.7178, and LAC >10.5455) was screened out comprising 13 nodes and 56 edges (Figure 4D). The size of the node represented the degree of the node in the interactive PPI network, with larger nodes and deeper color meaning higher degree values. The top nodes possessing higher degrees were regarded as hub genes, containing JUN (n = 40), TP53 (n = 38), MAPK1 (n = 38), RELA (n = 36), MYC (n = 28), and ESR1 (n = 24) (Supplementary Table S3). These target proteins were prepared to be docked with the main active ingredients analyzed earlier.
FIGURE 4
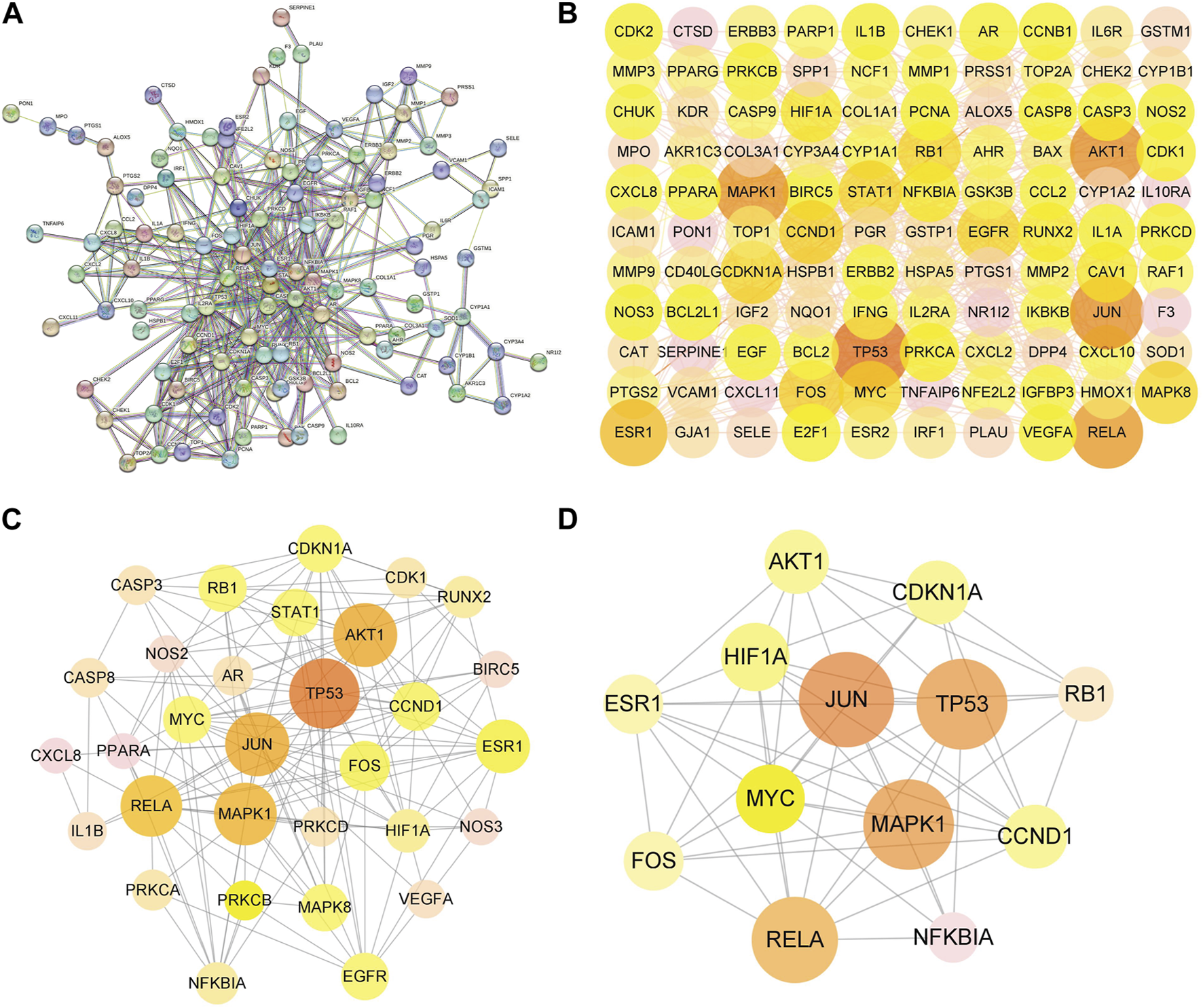
Protein–protein interaction (PPI) network of RPD against IBS and UC. (A) The interactive PPI network acquired from STRING database platform. (B) PPI network imported from STRING database to Cytoscape 3.8.0. (C) The PPI network of more significant proteins is composed of 31 nodes and 153 edges. (D) Core PPI network containing 13 nodes and 56 edges. A node with a larger size and deeper color possesses a higher degree value, such as JUN, TP53, MAPK1, RELA, and MYC.
GO biological processes of common target genes and KEGG pathway analysis
GO biological processes and KEGG pathway enrichment analysis were carried out to elucidate the functions and the enriched pathways of the potential treatment genes of RPD. The GO analysis consists of biological process (BP), cellular component (CC), and molecular function (MF). In this study, a total of 3,963 statistically significant GO terms were obtained with q-value <0.05, including 2053 of BP, 54 of CC, and 164 of MF. The top 10 terms were visualized in a bubble chart (Figure 5A). The redder the color of a dot, the lower the q-value and the greater the enrichment of the GO term. The results showed that response to oxidative stress (GO:0006979), response to xenobiotic stimulus (GO:0009410), and cellular response to chemical stress (GO:0062197) were tightly related to UC and IBS biological processes. The main disease-related terms in cellular components consisted of membrane raft (GO:0045121), membrane microdomain (GO:0098857), and transcription regulator complex (GO:0005667). Concerning molecular functions, more significant enrichment was found in DNA-binding transcription factor binding (GO:0140297), RNA polymerase II-specific DNA-binding transcription factor binding (GO:0061629), and ubiquitin-like protein ligase binding (GO:0044389).
FIGURE 5
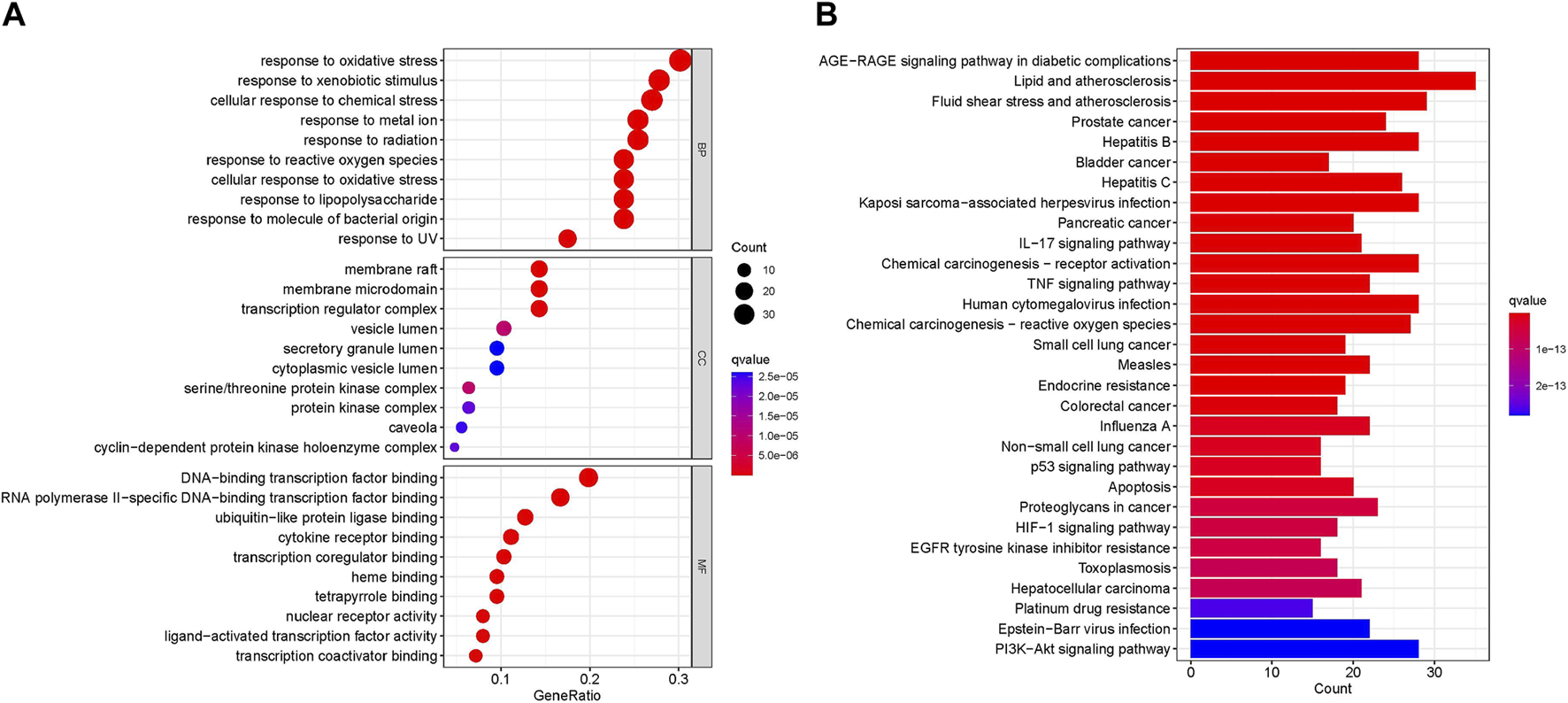
Enrichment Analysis (A) Bubble plot of GO enrichment analysis including the top 10 significant enrichment terms of three domains: BP, CC, and MF. The x-axis represents the gene proportion enriched in each entry, and y-axis shows the enrichment degree according to the q-value. (B) Barplot of KEGG enrichment pathways. The horizontal axis represents the number of target genes enriched in each entry, and the vertical axis shows the enrichment degree according to the corrected q-value. The redder bar means lower q value.
KEGG analysis concentrated on the anti-IBS and anti-UC signaling pathways of RPD. The results demonstrated 166 statistically significant relative pathways, the top 20 significant enrichment potential pathways with the highest gene counts were presented in a barplot diagram (Figure 5B), including “AGE-RAGE signaling pathway in diabetic complications,” “Lipid and atherosclerosis,” and “Fluid shear stress and atherosclerosis.” Additionally, five main pathways were involved in the treatment of common diseases by RPD: “NF-kappa B signaling pathway” (hsa04064), “MAPK signaling pathway” (hsa04010), “Inflammatory bowel disease” (hsa05321), and “Tp53 signaling pathway” (hsa04115) (Supplementary Table S4). Overall, these results indicated that RPD acts on the treatment of UC and IBS through multiple targets and multiple pathways involving inflammation, oxidative stress, and immunity. The KEGG map of MAPK and NF-kappa B signaling pathways is shown in Supplementary Figure S3.
Molecular docking analysis
To evaluate the affinity of the candidate bioactive compounds for related targets, molecular docking analysis was performed for mimicking the interaction between small ligand molecules and receptor protein macromolecules. The binding poses and interactions of five ingredients with six proteins were obtained with Autodock Vina v.1.2.2. Additionally, the binding energy between the two counterparts for each interaction was calculated to predict their affinity (Table 3). Docking results indicated 3D interactions between small ligand molecules (chemical compounds) and receptor protein macromolecules in PyMOL software. The heatmap showed that the docking of MYC and (+)-catechin had the lowest binding energy (−9.6 kcal/mol), indicating that their stability was higher than the other combinations (Figure 6). Some local structures of molecular docking in detail were illustrated in Figure 7. The key active ingredients of RPD can bind well with key target genes, including ESR1 ((+)-catechin) (Figure 7A), JUN (kaempferol) (Figure 7B), MAPK1 (beta-sitosterol) (Figure 7C), RELA (quercetin) (Figure 7D), TP53 (aloe-emodin) (Figure 7E), and MYC- (+)-catechin (Figure 7F). This shows that good binding activity might play an important role in the treatment of UC and IBS. The above docking results suggested that they exert significant antioxidant and anti-inflammatory effects in the function of RPD.
TABLE 3
| Ingredients structure | Binding energy (kcal/mol) | ||||||
|---|---|---|---|---|---|---|---|
| JUN | TP53 | MAPK1 | RELA | MYC | ESR1 | ||
| 1a02 | 1gzh | 6g54 | 2vge | 5i50 | 3os8 | ||
| Quercetin |
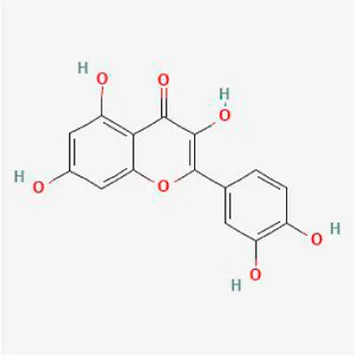
|
−8.2 | −7.2 | −8.5 | −6.8 | −8.5 | −8.3 |
| aloe-emodin |
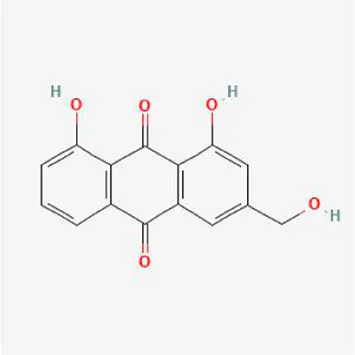
|
−9.4 | −7.2 | −8.7 | −6.7 | −8.9 | −8.4 |
| Kaempferol |
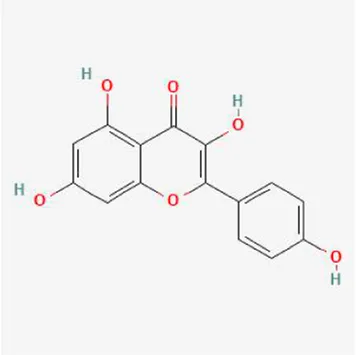
|
−9.4 | −7.0 | −8.4 | −6.8 | −8.3 | −8.4 |
| beta-sitosterol |
|
−7.4 | −7.1 | −8.7 | −7.0 | −7.9 | −7.5 |
| (+)-catechin |
|
−7.1 | −7.3 | −7.5 | −6.8 | −9.6 | −7.1 |
Docking results of five bioactive ingredients from RPD for IBS and UC targets.
FIGURE 6
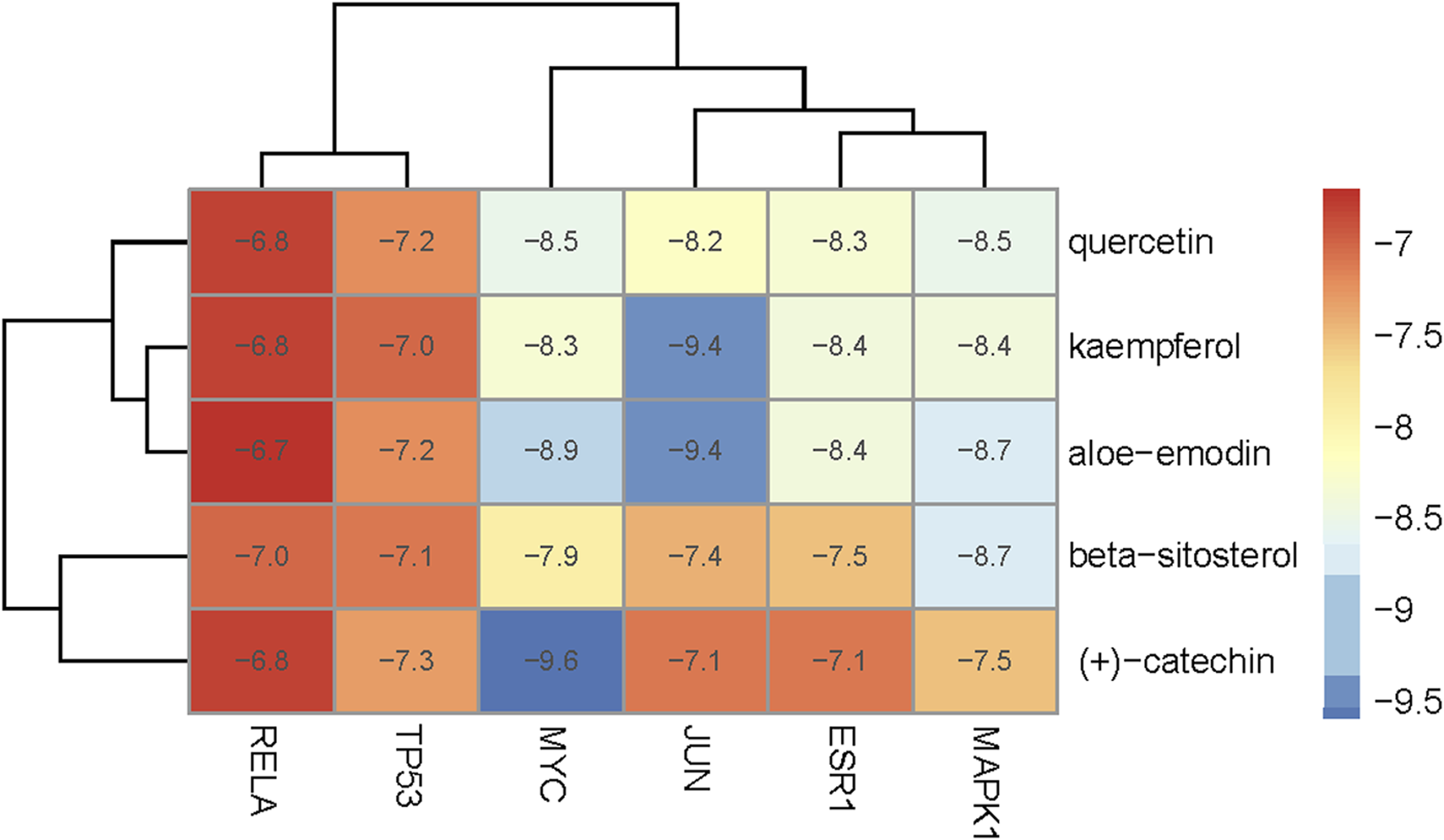
Heatmap based on binding energy among proteins and bioactives.
FIGURE 7
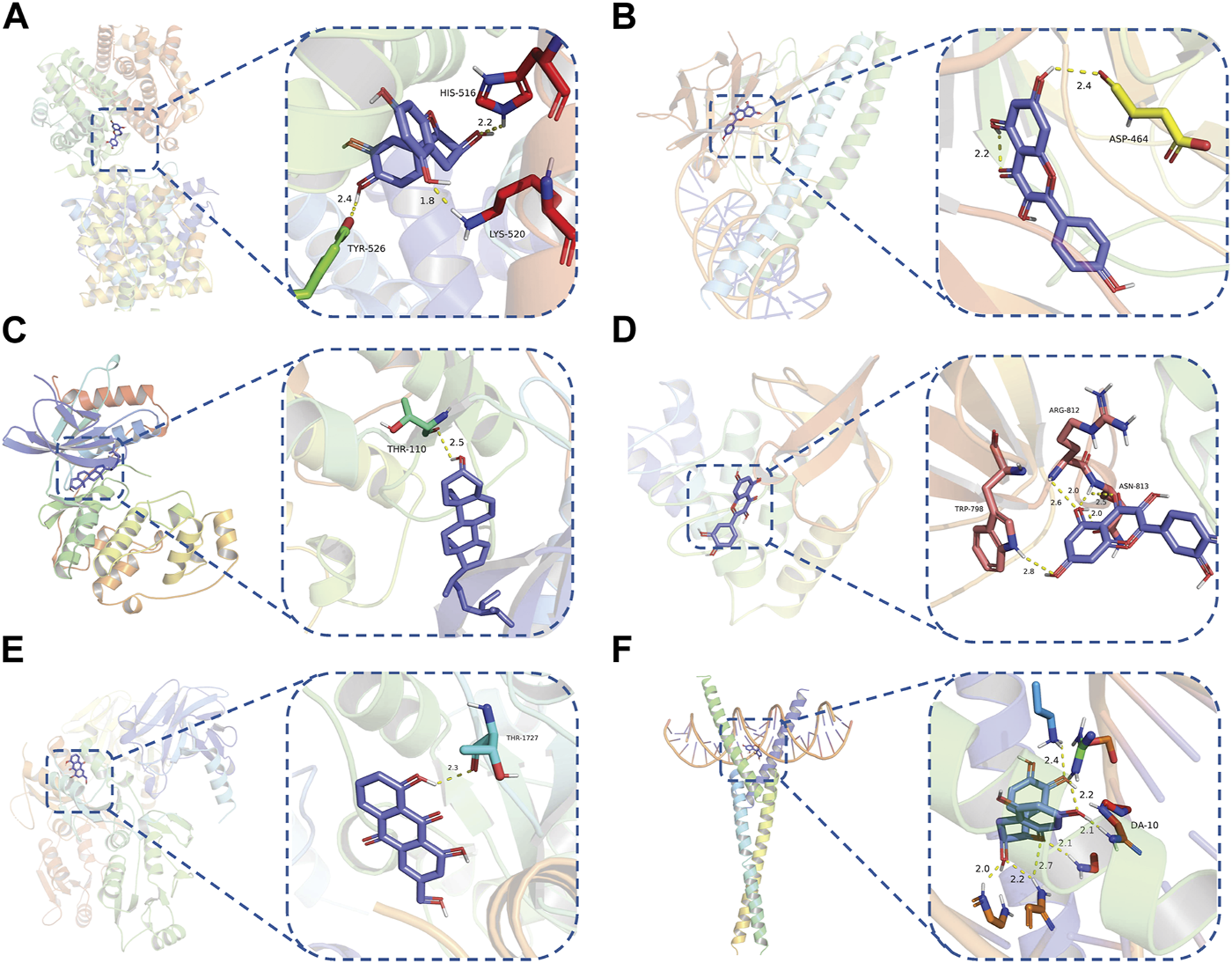
Binding mode of screened drugs to their targets by molecular docking: (A) ESR1- (+)-catechin; (B) JUN- kaempferol; (C) MAPK1-betasitosterol; (D) RELA- quercetin; (E) TP53- aloe-emodin; (F) MYC- (+)-catechin. Sticks represent ligands; the cartoon represents a hub target.
Discussion
Although the significant difference between UC and IBS is the endoscopic manifestation, more than 30% of quiescent UC patients report IBS-like symptoms, irrespective of whether they are accompanied by low-grade inflammation or not. UC concurrent IBS tends toward more severe psychological symptoms and worse quality of life, so proper intervention is warranted [25, 26]. Despite various established treatments for UC or IBS, promising feasible therapies for the overlapping symptoms remains a challenge. It still represents a therapeutic conundrum in the clinic because of the lack of comprehensive guidelines. As chronic intestinal inflammation exerts multi-system effects with heterogeneous symptoms and numerous complications, long-term western medicine treatment facesd problems such as gradual reduction of efficacy and many side effects, and more drug targets need to be explored.
Guided by a holistic view and syndrome differentiation, TCM achieves multi-target and multi-pathway efficacy through prescription and prescription coordination and interaction. Network pharmacology is an excellent tool to construct a drug-target network and reveal the underlying mechanisms of TCM. Spleen deficiency was regarded as the original cause of UC based on TCM theory [27]. Additionally, spleen deficiency and liver stagnation are the root causes of IBS [28, 29]. There is a certain internal relationship between the constitution and the syndrome, so the same treatment can be adopted. Nowadays, RPD has extensively been used for UC in previous studies, and its specific effectiveness was acknowledged in improving intestinal mucous lesions, modifying gut microbiota dysbiosis, and reducing inflammatory response [14, 30]. However, there is a lack of holistic and systematic understanding of the material basis and molecular pharmacological mechanism of its efficacy. Therefore, we performed a multi-level network to further clarify the underlying regulatory mechanism of RPD in the process of UC and IBS, thus enabling the provision ofadequate recommendations regarding potential treatment options.
By integrating all the targets of RPD and disease, 126 relative targets were identified, and top targets such as JUN, TP53, MAPK1, RELA, MYC, and ESR1 in the PPI network were considered as hub genes. Meanwhile, the GO and KEGG analysis revealed that inflammation, oxidative stress, immunity, oncogenicity, and gut microbiota dysbiosis are significantly involved in the pathological mechanisms of UC and IBS. BP enrichment results indicated that a variety of genes were associated with oxidative stress, including “response to oxidative stress,” “response to xenobiotic stimulus,” and “cellular response to chemical stress.” Emerging evidence has made great strides in clarifying that oxidative stress plays a crucial role in the development of colitis [31]. During inflammatory episodes, diffusive inflammatory cell infiltration and small intestinal mucosal crypt abscesses in colitis trigger the over-production of reactive oxygen species (ROS), followed by intestinal cell layer damage via free radical-dependent apoptosis. These pathological states lead to pathogen invasion and exaggeration of inflammatory cell infiltration as well as inflammatory damage [32], which ultimately promotes the initiation and progression of UC. Furthermore, several studies associate mild inflammation with IBS, and oxidative stress may be a crucial pathological factor since tissue damage due to oxidative attack is currently associated with inflammation [33, 34]. Moreover, an extensive study reported shreds of evidence of immune activation and epithelial barrier dysfunction in IBS patients with no other features of overt inflammation, containing intraepithelial lymphocyte aggregation and proinflammatory cytokine expression [35]. Therefore, ROS-targeted treatment and antioxidant prevention were proposed to be potential measures for alleviating UC and IBS [36].
Furthermore, the molecular docking results indicate that MAPK1, RELA, TP53, JUN, MYC, and ESR1, have superior affinities to the five core compounds quercetin, kaempferol, aloe-emodin, beta-sitosterol and (+)-catechin, which verified that colon damage may be repaired by improving inflammation, and reducing oxidative stress. Quercetin is a natural polyphenolic compound belonging to the flavonoid family. It is considered the most effective reactive oxygen species (ROS) scavenger by upregulating proinflammatory factors. Multiple animal models have confirmed quercetin exerts antioxidant and anti-inflammatory effects [37] by appearing to be mediated via interference in signal transduction pathways including the NF-κB and kinase systems [38]. Quercetin could offer dose-dependent protection for colitis mice through an antioxidant-like effect [39]. It is congruent with the fact that inflammatory responses are the important pathological mechanism of UC and IBS. Kaempferol is also a flavonoid with anti-inflammatory effects and has potential effects on alleviating murine experimental colitis by inhibiting the LPS-TLR4-NF-κB Axis and altering gut microbiota [40].
Likewise, aloe-emodin (a bioactive component of rhubarb), with a higher degree value in the PPI network, exerted an anti-inflammatory role by suppressing the secretion of inflammatory factors, downregulating the cascade reaction of NF-κB, MAPK, and PI3K pathways [41]. Aloe vera improved UC by enhancing colon mucus barrier functions in addition to reducing inflammation [42]. Increasing experimental and clinical evidence demonstrated that emodin exerted multi-targeting therapeutic mechanisms, involving anti-inflammatory, immunomodulatory, anti-fibrosis, and anti-tumor properties [15]. Catechins-flavanols attributes to flavonoids with diverse biological functions. They could exert potent antioxidant effects by inducing apoptosis, arresting cell-cycle, inhibiting NF-κB, and suppressing COX-2 overexpression in vitro and animal experiments [43]. β-Sitosterol is a phytosterol in plants that may play an anti-inflammatory role by reducing the production of proinflammatory cytokine and ROS [44]. Kang et al. found that novel effects of β-Sitosterol contributed to improving UC by exerting antimicrobial activity [45]. Taken together, the bioactive ingredients screened from RPD played significant roles in anti-inflammatory properties. These pharmacological activities indicated the interaction between “multiply compounds,” “multiply targets,” and “multiply pathways” of RPD.
KEGG analysis results demonstrated that the mechanism of RPD in the treatment of UC and IBS was mainly related to the following pathways indicating multiple targets and multiple pathways: “AGE-RAGE signaling pathway in diabetic complications,” “Inflammatory bowel disease,” “NF-κB signaling pathway,” “MAPK signaling pathway” and “Tp53 signaling pathway.” AGE-RAGE signaling pathway was involved in and mediated various signaling pathways by activating the PI3K-Akt and MAPK as well as the NF-κB signaling pathway, to induce ROS production and lead to inflammatory responses. NF-κB is acknowledged as a sign and the central pathway of inflammatory response, mediating various signaling pathways of inflammatory response [46]. The transcription factor NF-κB initiates expression of proinflammatory cytokines including IL-6, TNF-α, and IFN-γ [47]. The polymorphism of NFKB1A is associated with an increased risk of UC [48]. RELA is also a crucial member of the NF-κB family. Saito et. al claimed that compared to the resting proximal colon, a higher methylation level of ESR1 was detected in the actively inflamed colon [49]. Advanced glycosylation end products (AGE) and IL-17 can also be used as mediators for NF-κB pathway activation [50, 51]. Higher RAGE levels and RAGE polymorphisms have been found in UC patients correlated with the nuclear expression of pro-inflammatory, pro-oxidant, and pro-oncogenic genes [52], thus these pathways may be the potential inhibition point of intestinal symptoms and the progression to colorectal cancer (CRC) [47].
It is well-known that CRC is associated with the inflammation–dysplasia sequence, which poses a major threat to the prognosis of UC and IBS. The incidence of UC-related colon cancers was 1.6∼3.7%, accounting for 5.4% in all colon types [53, 54]. Chronic inflammation, the increased turnover of epithelial cells, and ROS production are thought to contribute to the development of dysplastic lesions, which may then transform into CRC [55]. JUN is a proto-oncogene that can induce oncogenic transformation [56]. The mutation of the p53 gene is a critical genetic change, involved in the early stages of UC-associated carcinogenesis of the colorectum. It is also used as a biomarker in predicting the risk of evolution toward malignancy [57].
Gut microbiota alternation is one of the underlying factors in perpetuating IBS symptoms [58]. Both our study and another study confirmed that the action mechanism of RPD is related to microbiota [14]. RPD could restore α diversity by enhancing Firmicutes and Actinobacteria and reducing Proteobacteria and Bacteroidetes [14]. Furthermore, quercetin was verified to exert a therapeutic effect in UC by altering the composition of gut microbiota [59]. The abundance of Bifidobacterium, Bacteroides, and Lactobacillus increased significantly while Fusobacterium and Enterococcus decreased totally. These findings indicate that RPD has the comprehensive ability to modify gut microbiota and suppress pro-inflammatory cytokines. Therefore, our results indicate that RPD may also have a role in regulating intestinal microbiota.
UC is a chronic recurrent intestinal inflammatory disease, whereas IBS is a functional intestinal disorder. The current clinical treatments for them are different, as IBS requires mainly symptomatic treatments. And the goal of UC treatment is to achieve clinical or endoscopic remission. It is challenging to tell the difference between IBS patients and UC in remission due to the numerous similarities between the two. Timely and accurate identification is key to avoiding overtreatment of IBS or delayed diagnosis of UC. When the two conditions are difficult to distinguish, Chinese medicine may be a proper option. However, the present study has several limitations. Firstly, absorption pathways, effective sites, and dose-effect outcomes, markedly effective components of bioactives in RPD, should be studied in depth. Secondly, the complex interaction between gut microbiota and herbal compounds needs more exploration. Moreover, any single certain bioactive detected in RPD is not sufficient to stand in completely for RPD. Thus, more experiments are required to further explore the potential molecular mechanism of RPD in the treatment of UC-IBS in vitro and in vivo.
Conclusion
In conclusion, both network pharmacology and molecular docking were applied to predict the common mechanism of RPD for the treatment of UC and IBS. The GO and KEGG pathways significantly enriched several signaling pathways involved in inflammation, oxidative stress, immunity, oncogenicity, and gut microbiota dysbiosis. Meanwhile, molecular docking results further verified that all the key active ingredients exhibited superior affinities to their core targets. RPD has revealed synergistic therapeutic effects in the treatment of UC and IBS partly by regulating multi-component, multi-target, and multi-pathway effects, which coincide with the biological mechanism of “treating different diseases with the same treatment.” This study offers novel insights into the effective therapeutic and management strategies for patients overlapping between UC and IBS, proposing future directions for research in this stimulating field.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
LZ and PY designed the research protocol. Study execution, acquisition of data, analysis and interpretation: LZ, WY, DL, and WZ. Drafting, revising or critically reviewing the article: LZ, WY, WZ, and MC. PY revised the paper. All authors made a significant contribution to the work reported, gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the fund from the Reginal Collaboration R&D Program of Sichuan Province (2022YFQ0053).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/jpps.2023.11225/full#supplementary-material
Abbreviations
IBS, Irritable Bowel Syndrome; UC, Ulcerative Colitis; IBD, Inflammatory bowel disease; RPD, Rhubarb peony decoction; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; PPI, Protein-Protein Interaction; MAPK, Mitogen-activated protein kinase; OB, Oral bioavailability; DL, Drug likeness.
References
1.
Ungaro R Mehandru S Allen PB Peyrin-Biroulet L Colombel JF . Ulcerative Colitis. The Lancet (2017) 389(10080):1756–70. 10.1016/s0140-6736(16)32126-2
2.
Ramos GP Papadakis KA . Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clinic Proc (2019) 94(1):155–65. 10.1016/j.mayocp.2018.09.013
3.
Ford AC Sperber AD Corsetti M Camilleri M . Irritable Bowel Syndrome. The Lancet (2020) 396(10263):1675–88. 10.1016/s0140-6736(20)31548-8
4.
Black CJ Ford AC . Global burden of Irritable Bowel Syndrome: Trends, Predictions and Risk Factors. Nat Rev Gastroenterol Hepatol (2020) 17(8):473–86. 10.1038/s41575-020-0286-8
5.
Alammar N Stein E . Irritable Bowel Syndrome: What Treatments Really Work. Med Clin North America (2019) 103(1):137–52. 10.1016/j.mcna.2018.08.006
6.
Lacy BE Moreau JC . Diarrhea-predominant Irritable Bowel Syndrome: Diagnosis, Etiology, and New Treatment Considerations. J Am Assoc Nurse Pract (2016) 28(7):393–404. 10.1002/2327-6924.12387
7.
Cangemi DJ Lacy BE . Management of Irritable Bowel Syndrome with Diarrhea: a Review of Nonpharmacological and Pharmacological Interventions. Therap Adv Gastroenterol (2019) 12:175628481987895. 10.1177/1756284819878950
8.
Dinan TG Quigley EM Ahmed SM Scully P O’Brien S O’Mahony L et al Hypothalamic-pituitary-gut axis Dysregulation in Irritable Bowel Syndrome: Plasma Cytokines as a Potential Biomarker? Gastroenterology (2006) 130(2):304–11. 10.1053/j.gastro.2005.11.033
9.
Koloski NA Jones M Kalantar J Weltman M Zaguirre J Talley NJ . The Brain-Ggut Pathway in Functional Gastrointestinal Disorders Is Bidirectional: a 12-year Prospective Population-Based Study. Gut (2012) 61(9):1284–90. 10.1136/gutjnl-2011-300474
10.
Matricon J Meleine M Gelot A Piche T Dapoigny M Muller E et al Review Article: Associations between Immune Activation, Intestinal Permeability and the Irritable Bowel Syndrome. Aliment Pharmacol Ther (2012) 36(11-12):1009–31. 10.1111/apt.12080
11.
Halpin SJ Ford AC . Prevalence of Symptoms Meeting Criteria for Irritable Bowel Syndrome in Inflammatory Bowel Disease: Systematic Review and Meta-Analysis. Am J Gastroenterol (2012) 107(10):1474–82. 10.1038/ajg.2012.260
12.
Spiller R Major G . IBS and IBD - Separate Entities or on a Spectrum?Nat Rev Gastroenterol Hepatol (2016) 13(10):613–21. 10.1038/nrgastro.2016.141
13.
Eichele DD Young R . Medical Management of Inflammatory Bowel Disease. Surg Clin North America (2019) 99(6):1223–35. 10.1016/j.suc.2019.08.011
14.
Luo S Wen R Wang Q Zhao Z Nong F Fu Y et al Rhubarb Peony Decoction Ameliorates Ulcerative Colitis in Mice by Regulating Gut Microbiota to Restoring Th17/Treg Balance. J Ethnopharmacology (2019) 231:39–49. 10.1016/j.jep.2018.08.033
15.
Zheng Q Li S Li X Liu R . Advances in the Study of Emodin: an Update on Pharmacological Properties and Mechanistic Basis. Chin Med (2021) 16(1):102. 10.1186/s13020-021-00509-z
16.
Ru J Li P Wang J Zhou W Li B Huang C et al TCMSP: a Database of Systems Pharmacology for Drug Discovery from Herbal Medicines. J Cheminform (2014) 6:13. 10.1186/1758-2946-6-13
17.
Liu Z Guo F Wang Y Li C Zhang X Li H et al BATMAN-TCM: a Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci Rep (2016) 6:21146. 10.1038/srep21146
18.
Xu HY Zhang YQ Liu ZM Chen T Lv CY Tang SH et al ETCM: an Encyclopaedia of Traditional Chinese Medicine. Nucleic Acids Res (2019) 47(D1):D976–82. 10.1093/nar/gky987
19.
Chen CY . TCM Database@Taiwan: the World's Largest Traditional Chinese Medicine Database for Drug Screening In Silico. PLoS One (2011) 6(1):e15939. 10.1371/journal.pone.0015939
20.
Szklarczyk D Gable AL Lyon D Junge A Wyder S Huerta-Cepas J et al STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–13. 10.1093/nar/gky1131
21.
Wang Y Bryant SH Cheng T Wang J Gindulyte A Shoemaker BA et al PubChem BioAssay: 2017 Update. Nucleic Acids Res (2017) 45(D1):D955–63. 10.1093/nar/gkw1118
22.
Morris GM Huey R Olson AJ . Using AutoDock for Ligand-Receptor Docking. Curr Protoc Bioinformatics (2008) 8:8.14. 10.1002/0471250953.bi0814s24
23.
Gaillard T . Evaluation of AutoDock and AutoDock Vina on the CASF-2013 Benchmark. J Chem Inf Model (2018) 58(8):1697–706. 10.1021/acs.jcim.8b00312
24.
Nong F Zhao Z Luo X Liu C Li H Liu Q et al Evaluation of the Influence of Mirabilite on the Absorption and Pharmacokinetics of the Ingredients in Dahuang-Mudan Decoction by a Validated UPLC/QTOF-MS/MS Method. Biomed Chromatogr (2019) 33(3):e4423. 10.1002/bmc.4423
25.
Berrill JW Green JT Hood K Campbell AK . Symptoms of Irritable Bowel Syndrome in Patients with Inflammatory Bowel Disease: Examining the Role of Sub-clinical Inflammation and the Impact on Clinical Assessment of Disease Activity. Aliment Pharmacol Ther (2013) 38(1):44–51. 10.1111/apt.12335
26.
Jelsness-Jørgensen LP Bernklev T Moum B . Calprotectin Is a Useful Tool in Distinguishing Coexisting Irritable Bowel-like Symptoms from that of Occult Inflammation Among Inflammatory Bowel Disease Patients in Remission. Gastroenterol Res Pract (2013) 2013:1–4. 10.1155/2013/620707
27.
Niu K Li Q Liu Y Qiao Y Li B Wei C et al Molecular Targets and Mechanisms of Scutellariae Radix-Coptidis Rhizoma Drug Pair for the Treatment of Ulcerative Colitis Based on Network Pharmacology and Molecular Docking. Evidence-Based Complement Altern Med (2021) 2021:1–15. 10.1155/2021/9929093
28.
Wang Y Fan H Qi X Lai Y Yan Z Li B et al Are Personalized Tongxie Formula Based on Diagnostic Analyses More Effective in Reducing IBS Symptoms?-A Randomized Controlled Trial. Complement Therapies Med (2018) 40:95–105. 10.1016/j.ctim.2018.07.002
29.
Zhang RZ Yu SJ Bai H Ning K . TCM-mesh: The Database and Analytical System for Network Pharmacology Analysis for TCM Preparations. Sci Rep (2017) 7(1):2821. 10.1038/s41598-017-03039-7
30.
Luo S Luo X Qi L Pan Z Zhou L . To Investigate the Therapeutic Effect and Mechanism of Rhein on DSS-Induced Ulcerative Colitis in Mice. Chin J Exp Formulary (2017) 23(11):109–13. 10.13422/j.cnki.syfjx.2017110109
31.
Moura FA de Andrade KQ Dos Santos JCF Araújo ORP Goulart MOF . Antioxidant Therapy for Treatment of Inflammatory Bowel Disease: Does it Work?Redox Biol (2015) 6:617–39. 10.1016/j.redox.2015.10.006
32.
Wang Z Li S Cao Y Tian X Zeng R Liao DF et al Oxidative Stress and Carbonyl Lesions in Ulcerative Colitis and Associated Colorectal Cancer. Oxidative Med Cell Longevity (2016) 2016:1–15. 10.1155/2016/9875298
33.
Achitei D Ciobica A Balan G Gologan E Stanciu C Stefanescu G . Different Profile of Peripheral Antioxidant Enzymes and Lipid Peroxidation in Active and Non-active Inflammatory Bowel Disease Patients. Dig Dis Sci (2013) 58(5):1244–9. 10.1007/s10620-012-2510-z
34.
Balmus IM Ciobica A Cojocariu R Luca AC Gorgan L . Irritable Bowel Syndrome and Neurological Deficiencies: Is There A Relationship? the Possible Relevance of the Oxidative Stress Status. Medicina (Kaunas). (2020) 56(4):175. 10.3390/medicina56040175
35.
Vivinus-Nébot M Frin-Mathy G Bzioueche H Dainese R Bernard G Anty R et al Functional Bowel Symptoms in Quiescent Inflammatory Bowel Diseases: Role of Epithelial Barrier Disruption and Low-Grade Inflammation. Gut (2014) 63(5):744–52. 10.1136/gutjnl-2012-304066
36.
Mitani T Yoshioka Y Furuyashiki T Yamashita Y Shirai Y Ashida H . Enzymatically Synthesized Glycogen Inhibits Colitis through Decreasing Oxidative Stress. Free Radic Biol Med (2017) 106:355–67. 10.1016/j.freeradbiomed.2017.02.048
37.
Sotnikova R Nosalova V Navarova J . Efficacy of Quercetin Derivatives in Prevention of Ulcerative Colitis in Rats. Interdiscip Toxicol (2013) 6(1):9–12. 10.2478/intox-2013-0002
38.
Tatiya-Aphiradee N Chatuphonprasert W Jarukamjorn K . Immune Response and Inflammatory Pathway of Ulcerative Colitis. J Basic Clin Physiol Pharmacol (2018) 30(1):1–10. 10.1515/jbcpp-2018-0036
39.
Dodda D Chhajed R Mishra J Padhy M . Targeting Oxidative Stress Attenuates Trinitrobenzene Sulphonic Acid Induced Inflammatory Bowel Disease like Symptoms in Rats: Role of Quercetin. Indian J Pharmacol (2014) 46(3):286–91. 10.4103/0253-7613.132160
40.
Qu Y Li X Xu F Zhao S Wu X Wang Y et al Kaempferol Alleviates Murine Experimental Colitis by Restoring Gut Microbiota and Inhibiting the LPS-TLR4-NF-Κb Axis. Front Immunol (2021) 12:679897. 10.3389/fimmu.2021.679897
41.
Hu B Zhang H Meng X Wang F Wang P . Aloe-emodin from Rhubarb (Rheum Rhabarbarum) Inhibits Lipopolysaccharide-Induced Inflammatory Responses in RAW264.7 Macrophages. J Ethnopharmacology (2014) 153(3):846–53. 10.1016/j.jep.2014.03.059
42.
Shi G Jiang H Feng J Zheng X Zhang D Jiang C et al Aloe Vera Mitigates Dextran Sulfate Sodium-Induced Rat Ulcerative Colitis by Potentiating colon Mucus Barrier. J Ethnopharmacology (2021) 279:114108. 10.1016/j.jep.2021.114108
43.
Bernatoniene J Kopustinskiene DM . The Role of Catechins in Cellular Responses to Oxidative Stress. Molecules (2018) 23(4):965. 10.3390/molecules23040965
44.
Liao PC Lai MH Hsu KP Kuo YH Chen J Tsai MC et al Identification of β-Sitosterol as In Vitro Anti-inflammatory Constituent in Moringa Oleifera. J Agric Food Chem (2018) 66(41):10748–59. 10.1021/acs.jafc.8b04555
45.
Ding K Tan YY Ding Y Fang Y Yang X Fang J et al β-Sitosterol Improves Experimental Colitis in Mice with a Target against Pathogenic Bacteria. J Cell Biochem (2019) 120(4):5687–94. 10.1002/jcb.27853
46.
Hoesel B Schmid JA . The Complexity of NF-Κb Signaling in Inflammation and Cancer. Mol Cancer (2013) 12:86. 10.1186/1476-4598-12-86
47.
Verstrepen L Bekaert T Chau TL Tavernier J Chariot A Beyaert R . TLR-4, IL-1R and TNF-R Signaling to NF-Κb: Variations on a Common Theme. Cell Mol Life Sci (2008) 65(19):2964–78. 10.1007/s00018-008-8064-8
48.
Bank S Skytt Andersen P Burisch J Pedersen N Roug S Galsgaard J et al Polymorphisms in the Inflammatory Pathway Genes TLR2, TLR4, TLR9, LY96, NFKBIA, NFKB1, TNFA, TNFRSF1A, IL6R, IL10, IL23R, PTPN22, and PPARG Are Associated with Susceptibility of Inflammatory Bowel Disease in a Danish Cohort. PLoS One (2014) 9(6):e98815. 10.1371/journal.pone.0098815
49.
Saito S Kato J Hiraoka S Horii J Suzuki H Higashi R et al DNA Methylation of colon Mucosa in Ulcerative Colitis Patients: Correlation with Inflammatory Status. Inflamm Bowel Dis (2011) 17(9):1955–65. 10.1002/ibd.21573
50.
Tóbon-Velasco JC Cuevas E Torres-Ramos MA . Receptor for AGEs (RAGE) as Mediator of NF-kB Pathway Activation in Neuroinflammation and Oxidative Stress. CNS Neurol Disord - Drug Targets (2014) 13(9):1615–26. 10.2174/1871527313666140806144831
51.
Zhang H Xia B Li J Zhao Q Chen Z Zhou R et al Expression and Clinical Significance of IL-17 and IL-17 Receptor in Ulcerative Colitis. J Huazhong Univ Sci Tech [Medical Sciences] (2016) 36(1):37–40. 10.1007/s11596-016-1538-2
52.
Moura FA Goulart MOF Campos SBG da Paz Martins AS . The Close Interplay of Nitro-Oxidative Stress, Advanced Glycation End Products and Inflammation in Inflammatory Bowel Diseases. Curr Med Chem (2020) 27(13):2059–76. 10.2174/0929867325666180904115633
53.
Eaden JA Abrams KR Mayberry JF . The Risk of Colorectal Cancer in Ulcerative Colitis: a Meta-Analysis. Gut (2001) 48(4):526–35. 10.1136/gut.48.4.526
54.
Jess T Rungoe C Peyrin-Biroulet L . Risk of Colorectal Cancer in Patients with Ulcerative Colitis: a Meta-Analysis of Population-Based Cohort Studies. Clin Gastroenterol Hepatol (2012) 10(6):639–45. 10.1016/j.cgh.2012.01.010
55.
Low END Mokhtar NM Wong Z Raja Ali RA . Colonic Mucosal Transcriptomic Changes in Patients with Long-Duration Ulcerative Colitis Revealed Colitis-Associated Cancer Pathways. J Crohn's Colitis (2019) 13(6):755–63. 10.1093/ecco-jcc/jjz002
56.
Hao L Alkry LT Alattar A Faheem M Alshaman R Shah FA et al Ibrutinib Attenuated DSS-Induced Ulcerative Colitis, Oxidative Stress, and Inflammatory cascade by Modulating PI3K/Akt and JNK/NFκB Pathways. Arch Med Sci (2022) 18(3):805–15. 10.5114/aoms/146792
57.
Kobayashi K Tomita H Shimizu M Tanaka T Suzui N Miyazaki T et al p53 Expression as a Diagnostic Biomarker in Ulcerative Colitis-Associated Cancer. Int J Mol Sci (2017) 18(6):1284. 10.3390/ijms18061284
58.
Canakis A Haroon M Weber HC . Irritable Bowel Syndrome and Gut Microbiota. Curr Opin Endocrinol Diabetes Obes (2020) 27(1):28–35. 10.1097/med.0000000000000523
59.
Lin R Piao M Song Y . Dietary Quercetin Increases Colonic Microbial Diversity and Attenuates Colitis Severity in Citrobacter Rodentium-Infected Mice. Front Microbiol (2019) 10:1092. 10.3389/fmicb.2019.01092
Summary
Keywords
ulcerative colitis, network pharmacology, molecular docking, irritable bowel syndrome, rhubarb peony decoction
Citation
Zhai L, Yang W, Li D, Zhou W, Cui M and Yao P (2023) Network pharmacology and molecular docking reveal the immunomodulatory mechanism of rhubarb peony decoction for the treatment of ulcerative colitis and irritable bowel syndrome. J. Pharm. Pharm. Sci 26:11225. doi: 10.3389/jpps.2023.11225
Received
28 January 2023
Accepted
28 April 2023
Published
25 May 2023
Volume
26 - 2023
Edited by
Fakhreddin Jamali, University of Alberta, Canada
Updates
Copyright
© 2023 Zhai, Yang, Li, Zhou, Cui and Yao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Yao, pingyaozh@sina.com
†ORCID: Leilei Zhai, orcid.org/0000-0002-6530-9530
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.