- Department of Pharmacy, Yangpu Hospital, School of Medicine, Tongji University, Shanghai, China
Purpose: Gastrointestinal perforation (GIP) is a fatal adverse event (AE). The AE of GIP induced by novel antineoplastic agents has attracted attention recently. We aimed to explore the AE signals of GIP related to novel antineoplastic agents comprehensively based on the FDA Adverse Event Reporting System (FAERS).
Methods: The FAERS database containing 71 quarters of records was used for analysis. Reporting odds ratio (ROR), information component (IC), and empirical Bayesian geometric mean (EBGM) were utilized to evaluate the signals of GIP associated with novel antineoplastic drugs. Standardization of drug names was by employing MedEx-UIMA software and Python. Data analysis and visualization were performed using MySQL Workbench and R software.
Results: After cleaning and handling the data, 5226 GIP cases were identified that were associated with new antineoplastic medications, where these agents were the main suspected contributors. A total of 37 novel antineoplastic drugs were detected with signals of GIP for ROR and IC. Only 22 drugs showed statistically significant signals for EBGM. We found the GIP signals of 22 novel antineoplastic drugs overlapped for the 3 indicators, including anti-vascular endothelial growth factor/vascular endothelial growth factor receptor, anti-endothelial growth factor receptor, immune checkpoint inhibitors, and so on.
Conclusion: The potential risk of GIP associated with several novel antineoplastic agents was identified through data mining, which provided valuable information on the safety risks associated with GIP among these drugs. The potential threat of GIP should be recognized and managed properly when using these novel antineoplastic agents.
Introduction
Gastrointestinal perforation (GIP) is a severe and relatively uncommon adverse event (AE), which is potentially mortal. Clinical attention has been drawn to GIP related to drugs owing to its serious outcome. The GIP may result from the use of drugs such as non-steroidal anti-inflammatory drugs, anticoagulants, corticosteroids, some antineoplastic agents, and some other drugs (1–4).
Developments and innovations in pharmaceutical industries have given rise to numerous new and effective anti-cancer treatments, such as small-molecule targeted drugs and monoclonal antibodies. Antibody-drug conjugates, programmed cell death protein 1 (PD-1)/programmed death ligand-1 (PD-L1), chimeric antigen receptor-T-cells (CRA-T), and so on that are rapidly emerging as a boon for oncology patients (5, 6). The gastrointestinal AE induced by the novel antineoplastic agents has been mentioned commonly, however, the AE of GIP associated with these new antineoplastic drugs is rarely reported. FDA added a black box and recommended permanent discontinuation of bevacizumab in patients with GIP (7). The inhibitors of vascular endothelial growth factor (VEGF) and its receptor (VEGFR) were potentially associated with GIP (8, 9). With more and more novel antineoplastic agents are available, it is of great clinical practical significance to systematically study the relationship between novel antineoplastic agents and GIP. Unfortunately, no large-scale and comprehensive research has confirmed this association yet.
The vast database of AE reports established by the U.S. FDA provides abundant real-world data on the safety of drugs of clinical applications of prescriptions (10). Most of all, it is a database that’s free and accessible to the public (11). Data mining is recognized as an excellent method for early detection of drug safety signals, which can be applied to predict and understand the safety of drugs (12). Pharmacovigilance research based on the combination of data mining methods with the FDA Adverse Event Reporting System (FAERS) database has become progressively prevalent in recent years. In our study, we used the FAERS database to provide reference evidence by calculating the AE signals with a view to a better understanding of the association between GIP and new antineoplastic agents for safe clinical use.
Methods
Data source
FAERS updates AE data quarterly. We retrieved all the data from the official website (11). Two kinds of data formats (ASCII and XML) are available, both of which are common and convenient to manage. The ASCII files for quarter 1 of 2004 (2004Q1) to quarter 3 of 2021 (2021Q3) were fetched. Each quarter file contains seven subfiles, providing information separately, as shown in Table 1.
Discrimination of objective drugs and AE
There are two fields in the DRUG files that relate to drug names, including the “drugname” and the “prod_ai.” Given that FAERS is a spontaneous reporting system (SRS), drug names may be documented as generic names, chemical structure names, trade names, synonyms, code names, abbreviations, and even incorrect names (13). Therefore, before data mining, all drug name records were standardized in the DEMO files as generic names. Firstly, the “drugname” and “prod_ai” fields were mapped to the specific RxNorm concepts, which contain a single active ingredient by using MedEx-UIMA software (MedEx-UIMA 1.3.8, Vanderbilt University, United States) (14). Secondly, we merged the fields containing compound ingredients after the first step of processing by using python 3.8 (Python Software Foundation, Wilmington, United States), and the pharmacist manually standardized the drug names. Finally, the data cleaning of the DRUG table was completed by screening and checking once again, by using the DrugBank database (15). The target drugs are novel antineoplastic agents, including but not limited to the following types of drugs: small-molecule targeted drugs as well as monoclonal antibodies. To minimize the probability of false positives, we selected only “role_cod” fields that play the role of “primary suspect (PS).”
The “pt” field in the REAC table is the name of AE written in the Medical Dictionary for Regulatory Activities (MedDRA) and represented as the “Preferred Terms” (PTs). The Standardised MedDRA (MedDRA® trademark is registered by ICH, version 24.0) queries (SMQs) was employed to access PTs related to GIP (16). A total of 24 PTs were identified as target AE terms after referring to books and literature, as described below: “Duodenal perforation,” “Duodenal ulcer perforation,” “Duodenal ulcer perforation, obstructive,” “Gastric perforation,” “Gastric ulcer perforation, obstructive,” “Gastrointestinal perforation,” “Ileal perforation,” “Ileal ulcer perforation,” “Intestinal perforation,” “Jejunal perforation,” “Jejunal ulcer perforation,” “Large intestine perforation,” “Oesophageal perforation,” “Peptic ulcer perforation,” “Rectal perforation,” “Small intestinal perforation,” “Oesophageal ulcer perforation,” “Large intestinal ulcer perforation,” “Small intestinal ulcer perforation,” “Intestinal ulcer perforation,” “Diverticular perforation,” “Gastrointestinal ulcer perforation,” “Upper gastrointestinal perforation,” “Lower gastrointestinal perforation.”
Statistical analysis
We mainly used three files, DEMO, DRUG, and REAC. According to FDA’s recommendations, data cleaning was first performed for the DEMO table. Above all, duplicate records were excluded. If the CASEID (the number used to identify FAERS cases) is the same, the latest FDA_DT (the date the FDA received the case) is picked. If the CASEID and FDA_DT are the same, the higher PRIMARYID (the unique value used to identify the reports) is selected.
Descriptive analysis was performed to describe information about the case, including gender, age, occupation of the reporter, and year of reporting. The following three indicators, including reporting odds ratio (ROR), information component (IC), and empirical Bayesian geometric mean (EBGM) were applied to measure the signals of GIP of the target drug (17). The 95% two-sided confidence interval (CI) was calculated. The ROR is based on frequentist and it is extensively utilized because it’s simple to calculate and understand (18, 19). ROR05 (the lower limit of the 95% two-sided CI of the ROR) > 1 and the number of cases≥2 is considered with statistically significant signal (19–21). The IC is calculated by the Bayesian confidence propagation neural network (BCPNN) algorithm that was developed by the Uppsala Monitoring Center (UMC) (22, 23). The IC025 (the lower limit of the 95% two-sided CI of the IC) > 0 is regarded as with statistically significant signal (22, 24). The EBGM is based on the multi-item gamma Poisson shrinker (MGPS) algorithm, EBGM05 (the lower limit of the 95% two-sided CI of the EBGM) > 2, and the number of cases ≥0 is considered with statistically significant signal (25, 26). BCPNN and MGPS are both based on Bayesian methods, which tend to be more conservative in simulation studies than other methods and detect less signal in real data examples. As a result, BCPNN and MGPS have lower sensitivity and better specificity (18, 27). The calculation formulas and judging criteria for the three indicators are summarized in Supplementary Tables S1, S2.
Results
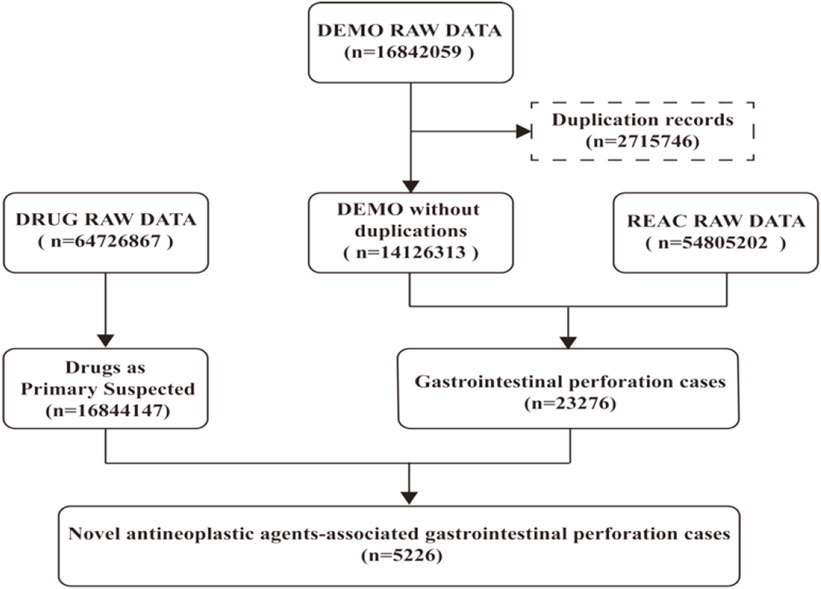
The DEMO files in the FAERS database documented 16842059 reports in the past 18 years, and 14126313 records were included in our study after deduplicating by following the FDA recommendations. 23276 cases were identified as GIP, of which 5,226 cases were judged being associated with the new antineoplastic medications (Figure 1). The clinical baseline data of the 5,226 cases was presented in Table 2. The sex percentage of reported cases were males (29.90%) verse females (26.90%). More than half of the patients were over 45 years old (63.61%).
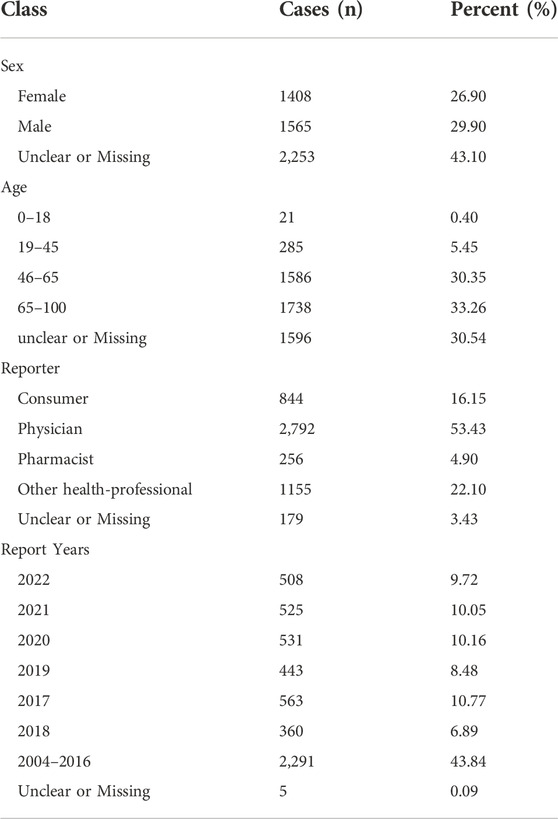
TABLE 2. Summary of patient characteristics with the AE of GIP related to novel antineoplastic agents.
A total of 37 novel antitumor drugs were detected with signals of GIP in terms of ROR and IC indexes (Figures 2, 3). Bevacizumab was identified with 13 signals that ranked the first, and the PT with the strongest signal was “gastrointestinal perforation” (ROR05 = 53.18, IC025 = 4.90). Both cetuximab and lenvatinib ranked the second concurrently, each with 10 statistically significant signals. For cetuximab, the strongest signal was “rectal perforation” (ROR05 = 4.90, IC025 = 1.47). The strongest signal of PT was “lower gastrointestinal perforation” for lenvatinib (ROR05 = 38.46, IC025 = 2.32). Atezolizumab was found with 8 signals, and the strongest signal of PT was “duodenal perforation” (ROR05 = 4.55, IC025 = 1. 58). In contrast, only 22 drugs presented statistically significant signals based on the EBGM (Figure 4). The detailed results were shown in Supplementary Table S3.
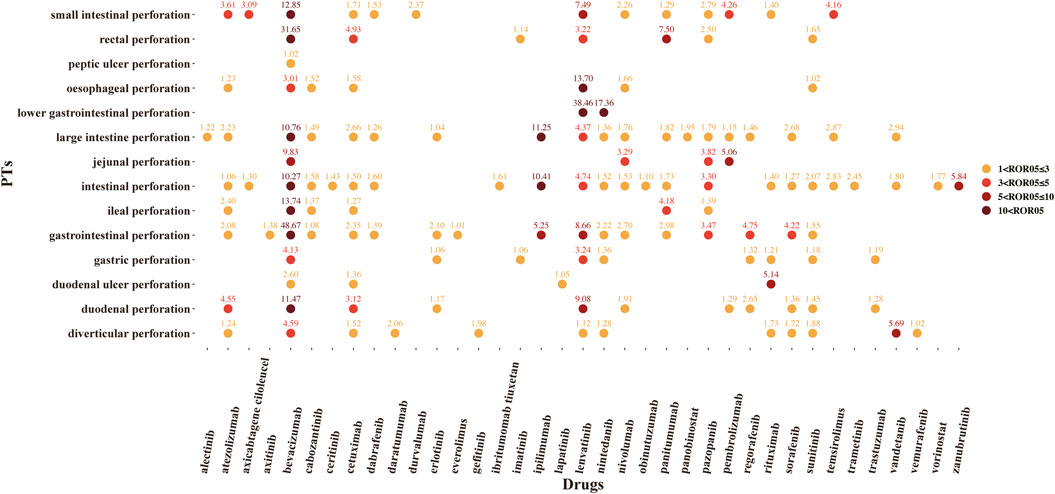
FIGURE 2. The signals of reporting odds ratio (ROR) of novel antineoplastic agents in detailed GIP AE PTs, preferred terms; ROR05, the lower end of the 95% confidence interval of ROR (ROR05 > 1 was regarded as the statistically significant signals).
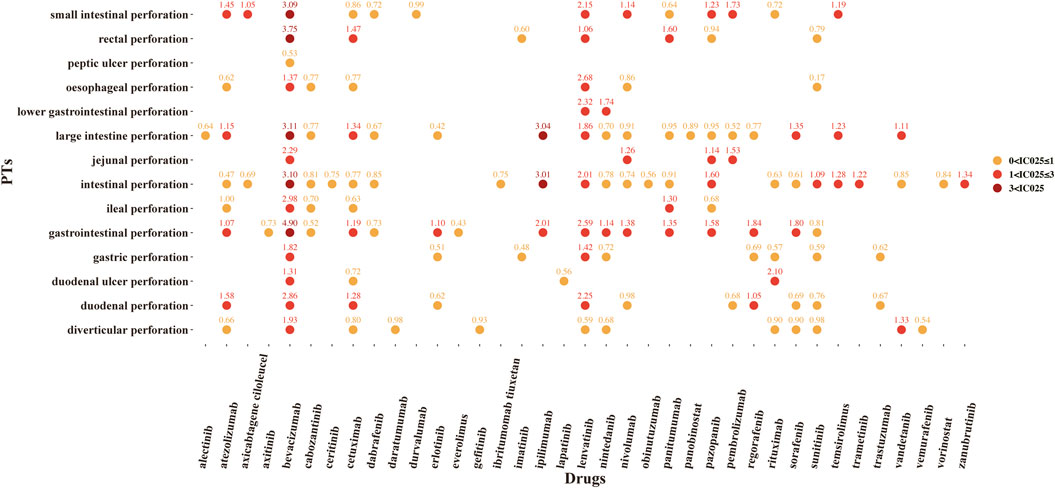
FIGURE 3. The signals of information component (IC) of novel antineoplastic drugs in detailed GIP AE PTs, preferred terms; IC025, the lower end of the 95% confidence interval of IC (IC025 > 0 was regarded as the statistically significant signals).
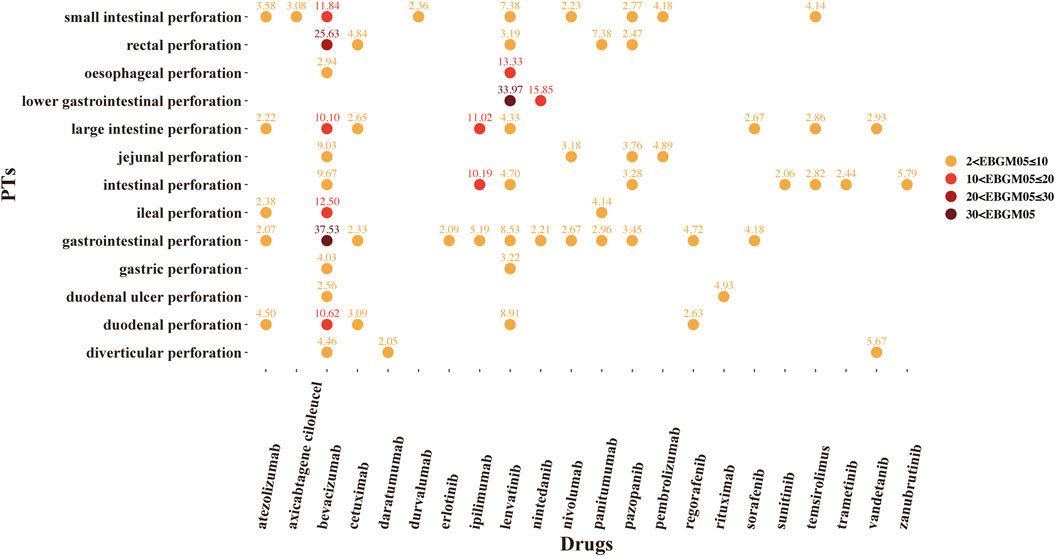
FIGURE 4. The signals of empirical Bayesian geometric mean (EBGM) of novel antineoplastic drugs in detailed GIP AE PTs, preferred terms; EBGM05, the lower end of the 95% confidence interval of EBGM (EBGM05 > 2 was regarded as the statistically significant signals).
There were 37 novel antitumor drugs being detected with signals of GIP based on ROR and IC indexes. Totally, for ROR, IC, and EBGM indicators, we found that there were 22 novel antineoplastic agents associated with overlapping GIP-related signals that including anti-VEGF/VEGFR, anti-endothelial growth factor receptor (EGFR), PD-1/PD-L1, and so on. The targets of the 22 drugs are summarized in Table 3.
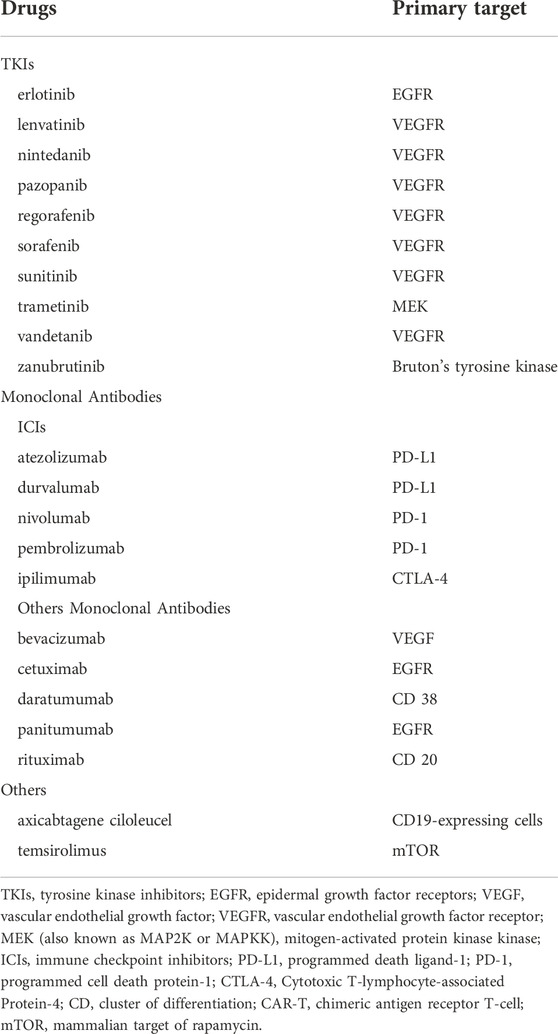
TABLE 3. Association of GIP signals with novel antineoplastic agents for the overlapped signals of the 3 indicators.
Discussion
The availability of novel antineoplastic drugs, especially for molecularly targeted drugs and tumor immune agents have dramatically changed the status of tumor treatment, and more and more patients benefit from these new treatments. However, these novel antineoplastic drugs have the risk of gastrointestinal perforation, a rare but fatal adverse event.
Bevacizumab was found to have the most potential to increase the risk of GIP, Wichelmann et al. performed a descriptive study of GIP with bevacizumab based on the FAERS database (9). A meta-analysis showed that there was no statistical difference in the risk of GIP between VEGFR-tyrosine kinase inhibitors (TKIs) and control groups (28). However, cases of GIP caused by anti-VEGF/VEGFR agents have been continuously reported in recent years (29–31). Wang Z et al. found that ramucirumab, a novel anticancer drug that belongs to the class of anti-VEGF agents, is associated with a significant increase in the risk of GIP (32). No comprehensive study has been conducted for comparative evaluation of the relationship of GIP associated with VEGF/VEGFR inhibitors. Data mining was applied to comprehensively estimate the GIP pharmacovigilance signal associated with the use of VEGF/VEGFR inhibitors. We found that VEGF/VEGFR inhibitors were associated with GIP, especially for bevacizumab, lenvatinib, and vandetanib.
The exact pathological mechanism by which VEGFR inhibitors cause perforation of the gastrointestinal tract remains unclear. However, the following inferences may be rational. Firstly, VEGF inhibition is thought to interfere with platelet-endothelial cell interactions, which may lead to loss of gastrointestinal vascular integrity and gastrointestinal submucosal inflammation (33). Secondly, these medications may reduce the blood delivery to the intestinal wall from normal blood vessels, which can lead to ischemia of the gastrointestinal mucosa (34). Finally, patients are immunocompromised on account of chemotherapy, which may cause dysbiosis of the intestinal flora and increase the risk of GIP (35).
Anti-EGFR drugs seems to be associated with GIP in our present study, including erlotinib, cetuximab and panitumumab. To date, there are fewer reports of anti-EGFR agents associated with GIP. Gass-Jégu F. et al. reported two cases of GIP after taking erlotinib treatment (36). Anti-EGFR drugs may downregulate VEGF expression, which leads to ischemia in gastrointestinal tract tissues (34, 37). However, the official drug label of cetuximab and panitumumab didn’t mention GIP. The relationship of anti-EGFR with GIP needs to be confirmed by further studies with different analysis tools.
In recent years, immune checkpoint inhibitors (ICIs) are the focus of research in the field of tumor immunotherapy. Inhibitors of PD-1/PD-L1 and cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) targets have demonstrated significant therapeutic benefits in clinical applications (38–41). Gastrointestinal AEs are the most common immune-related AEs (irAEs) with ICIs and the essential reason for the discontinuation of ICIs (42–44). The GIP signals associated with atezolizumab, durvalumab, nivolumab, pembrolizumab, and ipilimumab were detected by all three indicators. For official drug label, nivolumab reported with GIP, pembrolizumab and ipilimumab reported with GIP only in combination, atezolizumab, and durvalumab were not reported in the instructions. Although our study differs from the manual report, it is undeniable that ICIs cause a high proportion of immune enterocolitis, which has the potential risk of causing GIP (45, 46). GIP signals associated with ICIs were detected by using the VigiBase database that confirmed our research (47). More and more studies reported cases of GIP after taking PD-1 (48, 49). Therefore, these signals need to be paid more attention. Mechanisms of GIP toxicity associated with ICIs remain to be clarified, however, it is considered that T cells, antibodies and cytokines contribute to the development of irAEs (50–52).
The TKIs of trametinib and zanubrutinib were detected with the GIP signals. Trametinib is one of the mitogen-activated protein kinase kinase (MEK, also known as MAP2K or MAPKK) inhibitors. Mourad N. et al. found that patients suffered GIP with the treatment of MEK inhibitors (53). The monoclonal antibodies of rituximab also showed the GIP signals. The GIP of rituximab therapy is rare but there, and several case reports indicated a possible association between rituximab and GIP (54–56). Although rare, rituximab might be associated with colitis that can be severe enough to cause colon perforation (57). Noteworthy, axicabtagene ciloleucel, one of the CAR-T therapies, was detected with the potential GIP signals in our study. A post-marketing surveillance study showed the new potential GIP signal for axicabtagene ciloleucel (58). A clinical study of the late effects of CAR-T cells reported that about 1.8% of patients died from duodenal ulcer and gut perforation (59). Because CAR-T immunotherapy is a novel antineoplastic treatment, its relationship with GIP still needs to be validated with a larger amount of post-marketing clinical data. Nevertheless, it is of great significance to learn about their potential and fatal AE.
One point to note is that no GIP was reported in the official drug label for zanubrutinib, cetuximab, panitumumab, daratumumab, and axicabtagene ciloleucel, but we detected relevant signals. It may be caused by unknown confounding factors or the limitations of the search and analytical conduct of the study. Furthermore, the signals detected in our study could be suggestive of potential risks, and whether there is an association between GIP and these drugs needs to be evaluated and interpreted by combining official drug labels, expert opinion, clinical data, and other relevant factors. Therefore, caution is needed to interpret our results showing the association of these drugs with GIP. However, GIP is a very serious AE, and these signals should still raise our alert.
The FAERS database provides a tremendous amount of information on AE, but unavoidably, limitations remain in our study. Firstly, it is inevitable that the clinical information might be missing or unknown because the FAERS is based on the SRS. Secondly, the total number of patients using these drugs cannot be accessed, so the exact incidence is extremely difficult to be calculated. Thirdly, to avoid false positives results, we considered only ‘PS’ agents and did not take into account concomitant drugs. The relevant drug interaction studies will be conducted in the future. Last but not least, due to the presence of confounding factors, the incidence and risk of GIP associated with those agents need to be studied and evaluated in prospective studies.
Conclusion
The potential risk of GIP associated with several novel antineoplastic agents, including anti-VEGF/VEGFR agents, anti-EGFR drugs, ICIs, etc., was comprehensively investigated by data mining. Our present study would provide valuable information on the different safety risks associated with GIP among these drugs. GIP could lead to fatal consequences, therefore, the potential threat of GIP should be kept in mind during the use of these novel antineoplastic agents. And the potential risk of GIP needs to be recognized and managed properly.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
ZY and XL came up with the idea of this present study. XL designed the investigational protocol. HZ, HC, and LZ were focused on the standardization of drug names and data cleaning. ZY and XL performed the summary analysis of the data. All authors contributed to performing the study and writing this paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our deeply gratitude to Yaoyong Huang for providing technical support for our investigation.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/jpps.2023.11235/full#supplementary-material
References
1. Bjarnason, I, Scarpignato, C, Holmgren, E, Olszewski, M, Rainsford, KD, and Lanas, A. Mechanisms of damage to the gastrointestinal tract from nonsteroidal anti-inflammatory drugs. Gastroenterology (2018) 154(3):500–14. doi:10.1053/j.gastro.2017.10.049
2. Enokida, T, and Tahara, M. Management of VEGFR-targeted TKI for thyroid cancer. Cancers (Basel) (2021) 13(21):5536. doi:10.3390/cancers13215536
3. Gaio, M, Ferrajolo, C, Zinzi, A, Riccardi, C, Di Filippo, P, Carangelo, L, et al. Association of direct oral anticoagulants (DOACs) and warfarin with haemorrhagic risk by applying correspondence analysis to data from the Italian pharmacovigilance database - a case study. Front Pharmacol (2021) 12:790740. doi:10.3389/fphar.2021.790740
4. Narum, S, Westergren, T, and Klemp, M. Corticosteroids and risk of gastrointestinal bleeding: A systematic review and meta-analysis. BMJ Open (2014) 4(5):e004587. doi:10.1136/bmjopen-2013-004587
5. Olivier, T, Haslam, A, and Prasad, V. Anticancer drugs approved by the US food and drug administration from 2009 to 2020 according to their mechanism of action. JAMA Netw Open (2021) 4(12):e2138793. doi:10.1001/jamanetworkopen.2021.38793
6. Rajha, E, and Yeung, SCJ. Toxicities of novel antineoplastic therapies. In: KH Todd, JCR Thomas, and K Alagappan, editors. Oncologic emergency medicine: Principles and practice. Cham: Springer International Publishing (2021). p. 711–38.
7. Hapani, S, Chu, D, and Wu, S. Risk of gastrointestinal perforation in patients with cancer treated with bevacizumab: A meta-analysis. Lancet Oncol (2009) 10(6):559–68. doi:10.1016/s1470-2045(09)70112-3
8. Tsang, VHM. Management of treatment-related toxicities in advanced medullary thyroid cancer. Curr Opin Oncol (2019) 31(3):236–42. doi:10.1097/cco.0000000000000534
9. Wichelmann, TA, Abdulmujeeb, S, and Ehrenpreis, ED. Bevacizumab and gastrointestinal perforations: A review from the FDA adverse event reporting system (FAERS) database. Aliment Pharmacol Ther (2021) 54(10):1290–7. doi:10.1111/apt.16601
10. Kumar, A. The newly available FAERS public dashboard: Implications for health care professionals. Hosp Pharm (2019) 54(2):75–7. doi:10.1177/0018578718795271
11.Food and Drug Administration. FDA adverse event reporting system (FAERS) quarterly data extract files (2020). Available from: https://fis.fda.gov/extensions/FPD-QDE-FAERS/FPD-QDE-FAERS.html (Accessed Apr 15, 2022).
12. Bone, A, and Houck, K. The benefits of data mining. Elife (2017) 6:e30280. doi:10.7554/eLife.30280
13. Khaleel, MA, Khan, AH, Ghadzi, SMS, Adnan, AS, and Abdallah, QM. A standardized dataset of a spontaneous adverse event reporting system. Healthcare (Basel) (2022) 10(3):420. doi:10.3390/healthcare10030420
14. Jiang, M, Wu, Y, Shah, A, Priyanka, P, Denny, JC, and Xu, H. Extracting and standardizing medication information in clinical text - the MedEx-UIMA system. AMIA Jt Summits Transl Sci Proc (2014) 2014:37–42.
15. Wishart, DS, Feunang, YD, Guo, AC, Lo, EJ, Marcu, A, Grant, JR, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res (2018) 46:D1074–D1082. doi:10.1093/nar/gkx1037
16.International Council for Harmonisation. International council for harmonisation (1990). Available from: https://www.meddra.org/ (Accessed Apr 17, 2022).
17. Ni, M, Yin, XD, Hu, WJ, Zeng, N, Zhao, B, and Li, ZL. Stevens-johnson syndrome following vancomycin and linezolid: A real-world analysis of post-marketing surveillance data. Front Pharmacol (2022) 13:872854. doi:10.3389/fphar.2022.872854
18. Poluzzi, E, Raschi, E, Piccinni, C, and De Ponti, F. Data mining techniques in pharmacovigilance: Analysis of the publicly accessible FDA adverse event reporting system (AERS). London, UK: Data mining applications in engineering and medicine: IntechOpen (2012). p. 265–302.
19. van Puijenbroek, EP, Bate, A, Leufkens, HGM, Lindquist, M, Orre, R, and Egberts, ACG. A comparison of measures of disproportionality for signal detection in spontaneous reporting systems for adverse drug reactions. Pharmacoepidemiol Drug Saf (2002) 11(1):3–10. doi:10.1002/pds.668
20. Fukazawa, C, Hinomura, Y, Kaneko, M, and Narukawa, M. Significance of data mining in routine signal detection: Analysis based on the safety signals identified by the FDA. Pharmacoepidemiol Drug Saf (2018) 27(12):1402–8. doi:10.1002/pds.4672
21. Harpaz, R, DuMouchel, W, LePendu, P, Bauer-Mehren, A, Ryan, P, and Shah, NH. Performance of pharmacovigilance signal-detection algorithms for the FDA adverse event reporting system. Clin Pharmacol Ther (2013) 93(6):539–46. doi:10.1038/clpt.2013.24
22. Bate, A, Lindquist, M, Edwards, IR, Olsson, S, Orre, R, Lansner, A, et al. A Bayesian neural network method for adverse drug reaction signal generation. Eur J Clin Pharmacol (1998) 54(4):315–21. doi:10.1007/s002280050466
23. Nomura, K, Takahashi, K, Hinomura, Y, Kawaguchi, G, Matsushita, Y, Marui, H, et al. Effect of database profile variation on drug safety assessment: An analysis of spontaneous adverse event reports of Japanese cases. Drug Des Develop Ther (2015) 9:3031–41. doi:10.2147/dddt.S81998
24. Tada, K, Maruo, K, Isogawa, N, Yamaguchi, Y, and Gosho, M. Borrowing external information to improve Bayesian confidence propagation neural network. Eur J Clin Pharmacol (2020) 76(9):1311–9. doi:10.1007/s00228-020-02909-w
25. Dumouchel, W. Bayesian data mining in large frequency tables, with an application to the FDA spontaneous reporting system. The Am Statistician (1999) 53(3):177–90. doi:10.1080/00031305.1999.10474456
26. Szarfman, A, Machado, SG, and O'Neill, RT. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDA's spontaneous reports database. Drug Saf (2002) 25(6):381–92. doi:10.2165/00002018-200225060-00001
27. Park, G, Jung, H, Heo, SJ, and Jung, I. Comparison of data mining methods for the signal detection of adverse drug events with a hierarchical structure in postmarketing surveillance. Life (Basel) (2020) 10(8):138. doi:10.3390/life10080138
28. Qi, WX, Sun, YJ, Tang, LN, Shen, Z, and Yao, Y. Risk of gastrointestinal perforation in cancer patients treated with vascular endothelial growth factor receptor tyrosine kinase inhibitors: A systematic review and meta-analysis. Crit Rev Oncology/Hematology (2014) 89(3):394–403. doi:10.1016/j.critrevonc.2013.10.002
29. Date, E, Okamoto, K, Fumita, S, and Kaneda, H. Gastrointestinal perforation related to lenvatinib, an anti-angiogenic inhibitor that targets multiple receptor tyrosine kinases, in a patient with metastatic thyroid cancer. Invest New Drugs (2018) 36(2):350–3. doi:10.1007/s10637-017-0522-4
30. Ouchi, R, Okada, K, Usui, K, Kurata, N, Suzuki, S, Nagao, M, et al. Intestinal perforation in a patient with colon cancer during treatment with regorafenib: A case report and review of the literature. Tohoku J Exp Med (2021) 254(3):207–11. doi:10.1620/tjem.254.207
31. Yoshimoto, T, Yoshikawa, K, Higashijima, J, Miyatani, T, Tokunaga, T, Nishi, M, et al. Bevacizumab-associated intestinal perforation and perioperative complications in patients receiving bevacizumab. Ann Gastroenterol Surg (2020) 4(2):151–5. doi:10.1002/ags3.12312
32. Wang, Z, Zhang, J, Zhang, L, Liu, P, Xie, Y, and Zhou, Q. Risk of gastrointestinal perforation in cancer patients receiving ramucirumab: A meta-analysis of randomized controlled trials. J Chemother (2016) 28(4):328–34. doi:10.1179/1973947815y.0000000053
33. Verheul, HMW, and Pinedo, HM. Possible molecular mechanisms involved in the toxicity of angiogenesis inhibition. Nat Rev Cancer (2007) 7(6):475–85. doi:10.1038/nrc2152
34. Sliesoraitis, S, and Tawfik, B. Bevacizumab-induced bowel perforation. J Am Osteopath Assoc (2011) 111(7):437–41.
35. Abu-Hejleh, T, Mezhir, JJ, Goodheart, MJ, and Halfdanarson, TR. Incidence and management of gastrointestinal perforation from bevacizumab in advanced cancers. Curr Oncol Rep (2012) 14(4):277–84. doi:10.1007/s11912-012-0238-8
36. Gass-Jégu, F, Gschwend, A, Gairard-Dory, AC, Mennecier, B, Tebacher-Alt, M, Gourieux, B, et al. Gastrointestinal perforations in patients treated with erlotinib: A report of two cases with fatal outcome and literature review. Lung Cancer (2016) 99:76–8. doi:10.1016/j.lungcan.2016.06.012
37. Petit, AM, Rak, J, Hung, MC, Rockwell, P, Goldstein, N, Fendly, B, et al. Neutralizing antibodies against epidermal growth factor and ErbB-2/neu receptor tyrosine kinases down-regulate vascular endothelial growth factor production by tumor cells in vitro and in vivo: Angiogenic implications for signal transduction therapy of solid tumors. Am J Pathol (1997) 151(6):1523–30.
38. Cella, D, Grünwald, V, Nathan, P, Doan, J, Dastani, H, Taylor, F, et al. Quality of life in patients with advanced renal cell carcinoma given nivolumab versus everolimus in CheckMate 025: A randomised, open-label, phase 3 trial. Lancet Oncol (2016) 17(7):994–1003. doi:10.1016/s1470-2045(16)30125-5
39. Kwon, ED, Drake, CG, Scher, HI, Fizazi, K, Bossi, A, van den Eertwegh, AJM, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): A multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol (2014) 15(7):700–12. doi:10.1016/s1470-2045(14)70189-5
40. Reck, M, Rodríguez-Abreu, D, Robinson, AG, Hui, R, Csőszi, T, Fülöp, A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med (2016) 375(19):1823–33. doi:10.1056/NEJMoa1606774
41. van der Heijden, MS, Loriot, Y, Durán, I, Ravaud, A, Retz, M, Vogelzang, NJ, et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma: A long-term overall survival and safety update from the phase 3 IMvigor211 clinical trial. Eur Urol (2021) 80(1):7–11. doi:10.1016/j.eururo.2021.03.024
42. Khoja, L, Day, D, Wei-Wu Chen, T, Siu, LL, and Hansen, AR. Tumour- and class-specific patterns of immune-related adverse events of immune checkpoint inhibitors: A systematic review. Ann Oncol (2017) 28(10):2377–85. doi:10.1093/annonc/mdx286
43. Kröner, PT, Mody, K, and Farraye, FA. Immune checkpoint inhibitor-related luminal GI adverse events. Gastrointest Endosc (2019) 90(6):881–92. doi:10.1016/j.gie.2019.09.009
44. Sanz-Segura, P, García-Cámara, P, Fernández-Bonilla, E, Arbonés-Mainar, JM, and Bernal Monterde, V. Gastrointestinal and liver immune-related adverse effects induced by immune checkpoint inhibitors: A descriptive observational study. Gastroenterología y Hepatología (2021) 44(4):261–8. doi:10.1016/j.gastrohep.2020.07.009
45. Soularue, E, Lepage, P, Colombel, JF, Coutzac, C, Faleck, D, Marthey, L, et al. Enterocolitis due to immune checkpoint inhibitors: A systematic review. Gut (2018) 67(11):2056–67. doi:10.1136/gutjnl-2018-316948
46. Carbonnel, F, Soularue, E, Coutzac, C, Chaput, N, Mateus, C, Lepage, P, et al. Inflammatory bowel disease and cancer response due to anti-CTLA-4: Is it in the flora? Semin Immunopathol (2017) 39(3):327–31. doi:10.1007/s00281-016-0613-x
47. Huang, S, Bai, X, Fang, T, Guo, Y, Zheng, K, and Lin, X. Gastrointestinal toxicities associated with immune checkpoint inhibitors: A disproportionality analysis leveraging VigiBase, the WHO adverse drug reaction database. J Zhejiang Univ Sci B (2021) 22(2):156–64. doi:10.1631/jzus.B2000449
48. Cho, HJ, Kim, WR, Kim, JH, Kim, DH, Kim, DJ, and Kang, H. Colonic perforation after treatment with nivolumab in esophageal cancer: A case report. Ann Coloproctol (2021) 37(1):S39–S43. doi:10.3393/ac.2020.00213.0030
49. Dabek, RJ, Bizanti, A, Thomas, S, Kalla, A, and Hamdallah, I. Squamous cell carcinoma of the tonsil causing small bowel perforation in the setting of metastasis and treatment with programmed death-1 inhibitors: A case report. Cureus (2020) 12(9):e10739. doi:10.7759/cureus.10739
50. Byrne, EH, and Fisher, DE. Immune and molecular correlates in melanoma treated with immune checkpoint blockade. Cancer (2017) 123(S11):2143–53. doi:10.1002/cncr.30444
51. Chen, DS, and Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity (2013) 39(1):1–10. doi:10.1016/j.immuni.2013.07.012
52. Tarhini, AA, Zahoor, H, Lin, Y, Malhotra, U, Sander, C, Butterfield, LH, et al. Baseline circulating IL-17 predicts toxicity while TGF-β1 and IL-10 are prognostic of relapse in ipilimumab neoadjuvant therapy of melanoma. J ImmunoTherapy Cancer (2015) 3:39. doi:10.1186/s40425-015-0081-1
53. Mourad, N, Lourenço, N, Delyon, J, Eftekhari, P, Bertheau, P, Allayous, C, et al. Severe gastrointestinal toxicity of MEK inhibitors. Melanoma Res (2019) 29(5):556–9. doi:10.1097/cmr.0000000000000618
54. Kollmar, O, Becker, S, Schilling, MK, and Maurer, CA. Intestinal lymphoma perforations as a consequence of highly effective anti-CD20 antibody therapy. Transplantation (2002) 73(4):669–70. doi:10.1097/00007890-200202270-00038
55. Cornejo, A, Bohnenblust, M, Harris, C, and Abrahamian, GA. Intestinal perforation associated with rituximab therapy for post-transplant lymphoproliferative disorder after liver transplantation. Cancer Chemother Pharmacol (2009) 64(4):857–60. doi:10.1007/s00280-009-1062-1
56. Ram, R, Ben-Bassat, I, Shpilberg, O, Polliack, A, and Raanani, P. The late adverse events of rituximab therapy-rare but there. Leuk Lymphoma (2009) 50(7):1083–95. doi:10.1080/10428190902934944
57. Mallepally, N, Abu-Sbeih, H, Ahmed, O, Chen, E, Shafi, MA, Neelapu, SS, et al. Clinical features of rituximab-associated gastrointestinal toxicities. Am J Clin Oncol (2019) 42(6):539–45. doi:10.1097/coc.0000000000000553
58. Fusaroli, M, Isgrò, V, Cutroneo, PM, Ferrajolo, C, Cirillo, V, Del Bufalo, F, et al. Post-marketing surveillance of CAR-T-cell therapies: Analysis of the FDA adverse event reporting system (FAERS) database. Drug Saf (2022) 45(8):891–908. doi:10.1007/s40264-022-01194-z
Keywords: gastrointestinal perforation, novel antineoplastic agents, adverse event, real-world study, pharmacovigilance
Citation: Yu Z, Zhu H, Chen H, Zhu L and Liao X (2023) Gastrointestinal perforation associated with novel antineoplastic agents: A real-world study based on the FDA Adverse Event Reporting System. J. Pharm. Pharm. Sci 26:11235. doi: 10.3389/jpps.2023.11235
Received: 14 December 2022; Accepted: 26 January 2023;
Published: 15 February 2023.
Edited by:
Fakhreddin Jamali, University of Alberta, CanadaCopyright © 2023 Yu, Zhu, Chen, Zhu and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolan Liao, MTUyNTk2NzYyNjBAMTI2LmNvbQ==, eXV6aWNoZW5nQHRvbmdqaS5lZHUuY24=,
 Zicheng Yu
Zicheng Yu Xiaolan Liao
Xiaolan Liao