- 1Hematology-Oncology Department, Faculty of Medicine, Saint Joseph University, Beirut, Lebanon
- 2Department of Human Genetics, Gilbert and Rose-Marie Chagoury School of Medicine, Lebanese American University, Beirut, Lebanon
- 3Medical Genetics Unit, Faculty of Medicine, Saint Joseph University, Beirut, Lebanon
- 4Urology Department, Faculty of Medicine, Saint Joseph University, Beirut, Lebanon
Bladder cancer (BC) has been associated with genetic susceptibility. Single peptide polymorphisms (SNPs) can modulate BC susceptibility. A literature search was performed covering the period between January 2000 and October 2020. Overall, 334 articles were selected, reporting 455 SNPs located in 244 genes. The selected 455 SNPs were further investigated. All SNPs that were associated with smoking and environmental exposure were excluded from this study. A total of 197 genes and 343 SNPs were found to be associated with BC, among which 177 genes and 291 SNPs had congruent results across all available studies. These genes and SNPs were classified into eight different categories according to their function.
Introduction
Bladder cancer (BC), one of the most common cancers worldwide, is particularly prevalent in developed countries [1, 2]. Its global incidence was estimated to be equal to 3%, in 2020, according to GLOBOCAN [1, 2]. It is a complex disease that involves different risk factors, mainly smoking and occupational carcinogen exposure [3], in addition to genetic susceptibility [4]. In fact, the lifetime absolute risk (AR) of developing BC in 50 years-old white non-Hispanic never-smoker men is 1.9%, whereas the AR is 7.1% for current smokers among 50 years-old white non-Hispanic men [5]. Recent advances in DNA sequencing technologies, in particular next-generation sequencing approaches, have opened new horizons, allowing a better understanding of genetic triggers related to BC [6]. Several genomic alterations were found to be linked to this specific cancer, including gene rearrangements, amplifications, deletions, copy number variations, and point variants, including pathogenic and polymorphic variants that are also known as single nucleotide polymorphisms (SNPs) [7]. SNPs, which account for 90% of the human genome’s variability, are gaining much interest in the oncogenetics field since many of them have been shown to modulate cancer susceptibility by increasing or decreasing the risk of an individual developing cancer [8]. Among genes associated with cancer, we can cite genes regulating environmental carcinogen metabolism, DNA repair, or cell cycle pathways, all of which are involved in the development and/or progression of any type of cancer, including BC [9, 10].
Many studies have examined the association of SNPs with BC in different populations. This extensive literature review aims to list all SNPs that are significantly associated with BC and discuss their involvement in this type of cancer.
Methods
In order to gather all available information related to the possible association of various SNPs with BC, an extensive literature search was performed in the PubMed database covering the period between January 2000 and October 2020. The keywords “bladder cancer” combined with “genetic predisposition” were used with Boolean operators. Overall, 894 articles were selected. The titles and abstracts of these articles were assessed for eligibility before evaluating their contents. Articles that target hereditary cancer, epigenetic studies, and linkage analysis studies were not included. At the end, 334 articles were selected, reporting 455 SNPs located in 244 genes. Selected genes were then classified based on the same method used by [11]. The article selection process is summarized in Figure 1 as a PRISMA flow diagram.
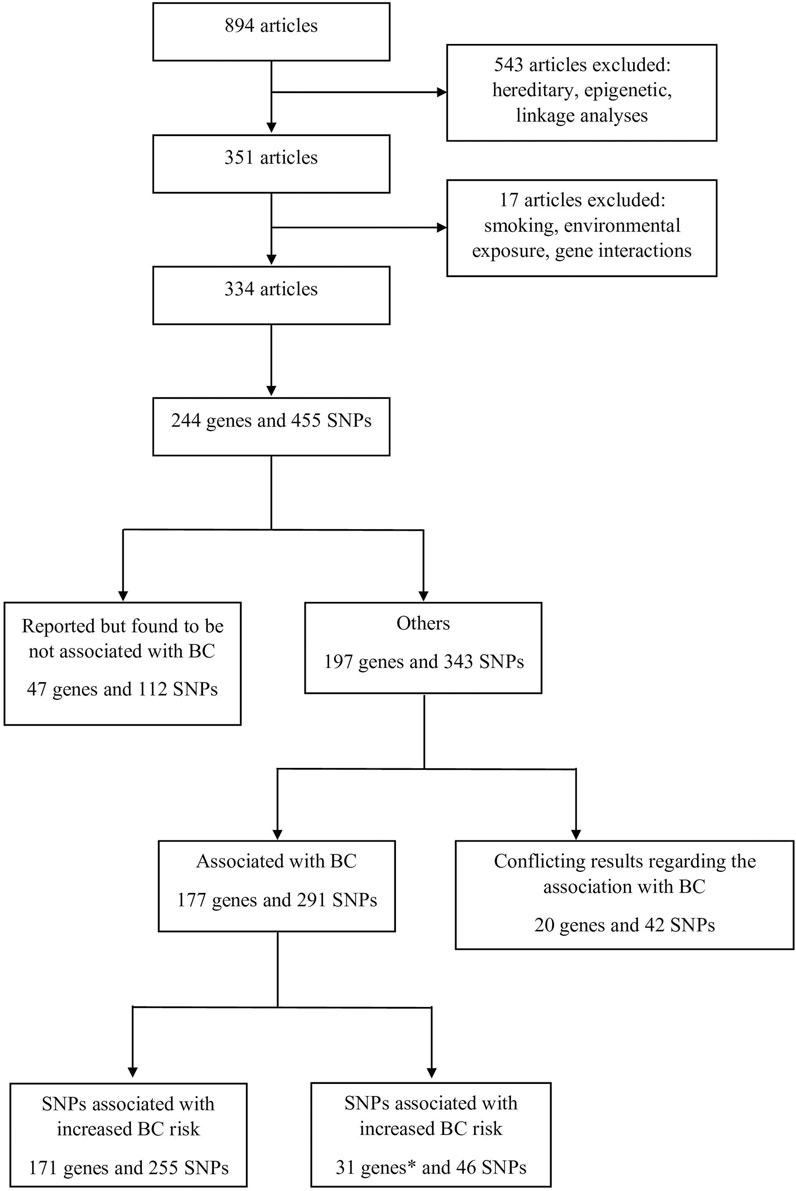
FIGURE 1. Process of the selection of the articles that were included in this study.* Genes with some SNPs that increase BC risk and others that decrease BC risk were counted twice.
The selected 455 SNPs were further investigated. All SNPs that were associated with smoking and environmental exposure but did not present a direct association with BC were excluded from this study. However, SNPs associated with BC independently of interfering factors like smoking status, sex, age, and others were included.
Results
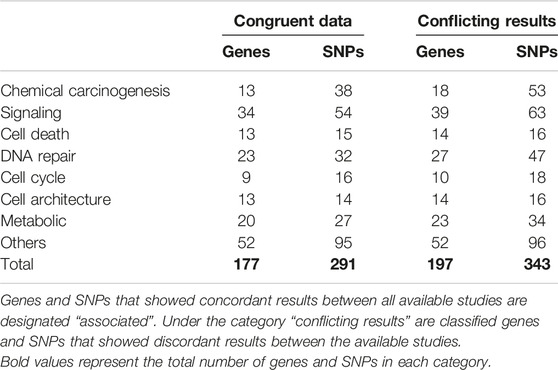
In total, 197 genes and 343 SNPs were found to be associated with BC, of which 177 genes and 291 SNPs had congruent results between all available studies, meaning that all studies that assessed a particular SNP had the same results regarding the SNP association with BC. The remaining 20 genes and 42 SNPs were reported with conflicting data, thus representing a huge amount of scattered data that needs a clear classification in order to facilitate their accessibility to researchers. These genes and SNPs were thus thoroughly evaluated and classified into eight different categories according to their function: chemical carcinogenesis, signaling, cell death, DNA repair, cell cycle, cell architecture, metabolic, and other genes that do not correspond to any specific category. Further subclassifications were also enclosed in each category (Table 1).
Chemical Carcinogenesis
Genetics and environmental factors impact the quantities of enzymes that participate in the activation and detoxification of chemicals, which can potentially lead to carcinogenesis. Most chemical compounds become carcinogens after being metabolized to chemically reactive electrophiles, which can interact with DNA to generate a carcinogenic response [12]. Genes encoding enzymes modulating chemical carcinogenesis were found to be linked to BC. In this section, genes were divided according to the different known enzyme families. Genes that encode for phase I and phase II enzymes were subcategorized into four categories: Cytochromes, Glutathione S-Transferase, N-acetyltransferase, and UDP-Glucuronosyltransferase (UGT).
Cytochromes are phase I enzymes whose role is to convert xenobiotics into excretable compounds. They are divided into subfamilies and are primarily located in the liver, but can also be found in the gastrointestinal tract, lungs, and kidneys. Mutations in these genes can affect substrate transformation, which may lead, in some cases, to cancer [13]. For instance, rs2472299 in CYP1A is known worldwide as a risk factor for BC [13], while rs4646903 and rs2198843 in the same gene [13, 14] are known to widely increase the risk for the same type of cancer but exclusively in the Asian population, and rs4646421 is specific for BC in Tunisians [15]. Results regarding rs1048943 in CYP1A1 are contradictory, since it increases the risk for BC in Asian and Brazilian populations but was not associated with BC in many other studies [13, 16–19]. Similarly, rs762551in CYP1A2 showed conflicting results [20–22]. Regarding the CYP1B1 gene: rs2855658 increases BC risks in the European population [23], and rs10012, rs1056827, and rs150799650 showed an increased risk in Indo-European cohorts [24], while rs1056827 was associated with a decreased risk in the Spanish population [25]. Contradictory results were seen for rs1056836, which presents a notable increased risk for BC in the Spanish population [13, 19, 25, 26]. On the other hand, rs4244285 and rs4986893 in CYP2C19 showed a protective effect against BC in the Chinese population [27]. CYP2E1 was associated with an increased risk for BC in the Lebanese population [28]. However, rs2031920 in the same gene was widely associated with a decreased risk in Asians [29] with controversial results in Caucasians [29, 30].
Glutathione S-Transferase constitutes a superfamily of phase II multipurpose enzymes that contribute to metabolic detoxification processes that protect macromolecules from environmental carcinogens, reactive oxygen species, and chemotherapeutic agents. The glutathione S-transferase family includes the following enzymes: GSTA, GSTM, GSTP, GSTT, and GSTO. Since these molecules contribute to the metabolism of potential carcinogens, any polymorphism affecting their expression or function may lead to cancer [31]. Most articles show that in most populations, the GSTM1 null genotype [32–47] and rs1695 in GSTP1 [16, 19, 33, 40, 42, 48–55] increase the risk for BC. However, a Chinese study showed that the GSTT1 null genotype was associated with a decreased risk for BC [36]. Rs156697 in GSTO2 also shows a positive association with BC in the Serbian population [56]. The polymorphisms in GSTT1 were mostly associated with an increased risk for BC [16, 18, 32–34, 40, 42, 44, 47, 48, 57–63]. Rs3957356, rs3957357, and rs4715332 in GSTA1 were associated with an increased risk for BC in both Balkan-Ben and Balkan-non Ben [63].
N-acetyltransferase (NAT) enzymes, also known as NAT1 and NAT2, are responsible for acetylating aromatic and heterocyclic amines in the liver, the gastrointestinal tract, and the urinary bladder. Genetic polymorphisms in these enzymes can alter the processing rate of various carcinogenic compounds, thus increasing the risk for cancer. Hence, slow NAT2 acetylation is associated with increased risk for BC [64]. Many SNPs in NAT1 are associated with a significantly increased risk for BC in the Lebanese population, including rs15561, rs4986782, and rs1057126 [28, 65, 66]. However, the latter did not show any association with BC in two other meta-analyses [4, 67]. Rs9650592 in NAT1 was positively associated with BC in the European population [23]. A study conducted on a French Caucasian population demonstrated an increased risk for BC in the presence of rs1208, rs1801280, or rs1041983 in NAT2 [19]. Rs1041983 presents the same effect in the American population [40]. Rs1799929, rs1799930, and rs1799931 were shown to be associated with BC in French and Bangladeshi populations [19, 68]. However, a Chinese meta-analysis contradicted the French results and found no association between these SNPs and BC [69]. Finally, rs1495741 and rs4646249 increase the risk of BC [11, 37, 70] in the European population [23].
The UDP-Glucuronosyltransferase (UGT) family of enzymes are phase II enzymes that are involved in the glucuronidation of aromatic amines and other carcinogens. They are primarily located in the liver but also in the gastrointestinal tract, lungs, and kidneys. The UGT family is composed of UGT1A, UGT2A, and UGT2B. They present different subtypes that are all, except for UGT2B17, expressed in normal bladder tissue [71]. Conflicting data was documented regarding the association of rs11892031 in UGT1A10/UGT1A8 with BC [23, 39, 70, 72–75]. Rs17863783 in UGT1A6 decreases the risk of developing BC [70, 76]. Rs1104892, rs1105880, rs1113193, rs1604144, rs17854828, rs17864684, rs17868322, rs2602374, rs2741042, rs2741044, rs2741045, and rs4148326 in UGT1A8 are associated with an increased risk in the American population, while rs1113193, rs1604144, rs17854828, rs17864684, rs4148328, rs4233633, and rs7571337 are associated with a decreased risk in the same population [77]. Rs2736520 in UGT2B4 increases the risk for BC, while Rs3822179 decreases the risk in the American population [77].
CDC-like kinase 3 (CLK3) is a dual specificity kinase that belongs to the serine/threonine type kinase family. Its role in human cancer is still undetermined [78]. However, a Japanese study has established that rs11543198 is associated with an increased risk for BC in the Japanese population [79].
Signaling
Signaling and immune-related genes were found to be associated with BC in the literature. These genes were divided into four categories, including cytokines, toll-like receptors, transcriptional factors, and other genes.
For an effective immune response against cancer cells, a sequence of steps must take place in fighting tumor cells. Each step of this sequence requires the presence of several stimulatory and inhibitory factors [80]. Among these actors, Cluster of Differentiation (CD) 274 [also known as Programmed Death Ligand 1 (PD-L1)], has an inhibitory function toward immune response activation. Rs4143815 in this gene increases the risk for BC in the Chinese population [81]. Cytotoxic T-lymphocyte-associated protein 4 (CTLA4) can also inhibit T cell-mediated immune responses, and rs231775 and rs3087243 in the corresponding gene have been shown to increase the risk for BC in the Indian population [82]. Intercellular Adhesion Molecule 1 (ICAM1) is responsible for T cell tumor infiltration, and therefore a variation in this gene can increase cancer susceptibility, as shown for rs5498 in the Taiwanese population [83]. Furthermore, chemokines help regulate the immune response and impact cancer progression [84]. Among the genes encoding chemokines, C-C chemokine receptor type 2 (CCR2) rs1799864 [85–87] and CCR5 rs333 [85] were associated with an increased risk for BC in the Turkish population, rs1801157 in C-X-C motif chemokine 12 (CXCL12) also has a positive association with BC in Indian [88] and Turkish [85] populations; and rs1126579 in CXC chemokine receptors 2 (CXCR2) increases cancer’s susceptibility in the Indian population [88] while it shows a protective effect towards BC in the American population [89]. Finally, rs187115 in CD44 was associated with an increased risk for BC in the Taiwanese population [90]. Tumor-secreted T-helper 17 (Th17) cell-associated interleukins (IL) mobilize immune suppressive cells and promote tumor growth [91]. Rs2275913 in IL-17A and rs187238 in IL-18 increase the risk for BC in the Polish population, with the highest association for IL-18 [92]. Rs1946518 in IL-18 also increases the risk of BC in the Indian population [93]. Rs2227485 in IL-22 is also a risk factor for BC in the Chinese population [94]. Other interleukins that play a role in macrophages and neutrophils’ immunosuppressive roles include IL-10, IL-12, and IL-23 [95]. Rs1800896, rs1800871, and rs1800872 in IL-10 [96] are associated with an increased risk for BC in Chinese [97] and Indian [98] populations; rs10889677 in IL-23R also increases BC risk in Chinese [99] and Polish [92] populations; and IL-12 decreases the risk in the Indian population [93]. Other interleukins were also found to play a role in BC risk; for instance, rs2069762 in the IL-2 gene increases the risk for BC in the Chinese population. Rs4073 in IL-8/CXCL8 gives high BC susceptibility in the Indian population [100] and rs1800890 in IL-19 in the Spanish population [101]. Last but not least, rs153109 and rs17855750 in IL-27 predispose to BC in Spanish and Chinese individuals, respectively [102, 103]. Finally, rs1799964 in TNF-α [104] and rs1800470 in TGFB1 [105] are positively associated with BC in the Indian population.
Toll-like receptors (TLR) play a key role in the initiation of the innate immune response. They are expressed on both immune and tumor cells and regulate the immune responses in tumor progression and as therapeutic targets for cancer [106]. TLR2 mutations (−196 to −174del) increase the risk of BC in India [107]. Rs11536889 in TLR4 [108–110] and rs72552316 in TLR7 [111] constitute separate BC risk factors.
Transcription factors might be linked to BC [112]. For instance, rs2228570 in VDR (Vitamin D receptor) was found to increase the risk of developing BC in India [113] and Tunisia [114]. TP63 can act as a tumor suppressor gene or as an oncogene depending on the cellular setting and pathways where it is implicated, and it can thus regulate the transcription of different genes [115]. Several SNPs have been shown to increase the risk of BC, among them rs4687100 [116] and rs710521 [39, 70, 116]. Of note, the latter has also been proven to present a protective effect in the Indian population [117], but no association with BC was identified in a Chinese study [118]. Finally, conflicting data were also obtained for rs35592567 since it was shown to have a positive association in one meta-analysis [11] and a negative association in another [119].
Vascular endothelial growth factor (VEGF) regulates angiogenesis and is upregulated and overexpressed in BC [120]. Rs699947 and Rs35569394 in VEGF increase and decrease, respectively, the BC risk in the Indian population [121]. Moreover, rs833052, rs25648 [122], rs3025039 [122, 123], and rs699947 [124] in VEGFA showed a positive association with BC, while the latter also presented a protective effect in the Tunisian population [125]. Rs3775194, rs1485762, rs6828869, and rs17697515 in VEGFC, and rs4557213 in VEGFR increase the risk for BC in the American Caucasian population [126] while rs1485766 in VEGFC increases the risk in the Taiwanese population [127]. Human Leukocyte Antigen G (HLA-G) expression in tumors promotes the immune suppressive microenvironment, which results in poor treatment response and prognosis [128]. Rs1063320, rs1610696, rs1704, rs1707, rs1710, rs17179101, and rs17179108 in HLA-G are associated with increased cancer susceptibility in the Brazilian population [129].
Other SNPs that were also studied in various genes include: rs6593205 and rs7799627 in Epidermal Growth Factor Receptor (EGFR), which decrease the risk for BC in the American Caucasian population, while rs11238349 increases the risk in the same population [126] and rs1050171 increases the risk in the Chinese population [130]. Rs696 in NFKBIA and Rs11188660 in BLNK (B cell linker) increase the risk in the Spanish population [101]. Rs28362491 in NFKB1 [131–133] and rs7944701 in MAML2 [134] increase the risk in the Chinese population. A more extensive list is available in Supplementary Table S1 under the section titled “signaling.”
Cell Death
By evading death, immortal cells can develop into tumors. Mutations in certain pathways promoting cell death can thus lead to tumor formation. Cell death pathways involve multiple genes, including cell-death receptors such as Tumor Necrosis Factor Receptor 1 (TNFR1), FAS, and TNF-related apoptosis-inducing ligand (TRAIL) receptors and effectors such as caspases [135]. Genes and SNPs related to those pathways were reported in BC. Rs2234767 in FAS, Rs763110 in FASLG [136], and Rs1131580 in TNFSF10 [137] increase the risk for BC in the Turkish population. In DR4 (TRAILR1 or TNFRSF10A), rs6557634 increases the risk for BC the Indian population [138] and rs13278062 increases the risk in the Chinese population [139] while rs20575 was found to present a protective effect in the Caucasian population [140]. Moreover, rs2647396 in BCL10 [101], rs10999426 in PRF1 [101], and rs42490 in RIPK2 [141] each individually increase the risk for BC in the Spanish population, while rs401681 in CLPTM1L increases the risk widely in all studied populations [39, 73, 74]. Rs4647603 in CASP3 and rs3181320 and rs507879 in CASP5 increase the risk for BC in Indians [138] while rs3834129 in CASP8 [142] and rs4645978 in CASP9 [143] decrease the risk for BC in the Chinese and Indian populations, respectively. Finally, rs1045411 in HMGB1 presents a protective effect in the Chinese population [144].
DNA Repair
The DNA repair genes associated with BC are classified according to the repair mechanism: base excision repair (BER), nucleotide excision repair (NER), homologous recombination (HR), and poly-ADP-ribosylation.
BER is a mechanism of DNA reparation that is not restricted to the repair of single-strand breaks but also of damages resulting from defects in alkylation, oxidation, deamination, and depurination. Given that this mechanism repairs thousands of errors per cell and per day, it plays a major role in cancer prevention by ensuring genome integrity and stability. Therefore, BER guarantees the integrality of apoptosis pathways and prevents mutation accumulation that may initiate tumors [145]. In X-ray repair cross-complementing protein 1 (XRCC1), rs1799782 and rs25489 are widely associated with an increased risk for BC, especially in Asian and Indian populations [146–152]. Rs915927 in the same gene increases tumor susceptibility in the Italian population [153] while rs25487 gave contradictory results in different studies [147–149, 154–159]. Rs3218373, rs3218536, and rs6464268 in XRCC2 present a protective effect towards BC in the Italian population [160]. Rs1805377 in XRCC4 increases the risk of BC in Spanish [160] and Indian individuals [161]. Rs6869366 increases the risk of BC in Taiwanese people [162] but decreases the risk in the Indian population [161]. Rs828907 in XRCC5 increases BC risk in the Taiwanese population [163]. Rs3213245 and rs7003908 in XRCC7 (PRKDC) are positively associated with BC, respectively, in the Chinese Han [164] and in the Indians [165]. Other SNPs in BC-associated BER genes include rs1760944 in APEX1, which decreases the risk for BC in the Chinese population [146] while rs1130409 in APE1 showed conflicting data [147, 166, 167]. Rs3136717 in POLB [168] and rs1052133 in hOGG1 [147, 165, 169–171] are associated with a high risk for BC, while rs125701 in hOGG1 decreases the risk for BC in the Spanish population [168]. Finally, rs2029167 in SMUG1 and rs3219487 in MUTYH increase the risk for BC in the American population [172] and rs11039130 in DDB2 has the same effect in the Caucasian population [173].
NER is also an important DNA repair pathway consisting of two distinct sub-pathways. The global-genome NER process fixes helix damages over the entire genome, whereas the transcription-coupled NER mechanism acts during transcription to resolve RNA polymerase blocking lesions [174]. Mutations in genes implicated in NER can result in a predisposition to cancer since mutations and chromosomal abnormalities can either activate oncogenes or inactivate tumor suppressor genes [175]. Rs3212961 in ERCC1 increases BC risk in the Spanish population [176] whereas rs967591, rs735482, and rs2336219 have a protective effect towards BC in the Italian population [153]. Rs13181 [15, 157, 177–182], rs1799793 [166, 170, 177, 178, 183–186] and rs238406 [176, 178, 179] in ERCC2 (XPD) were individually widely associated with an increased BC risk. Rs1047769 [176] and Rs17655 [187] in ERCC5 increase the risk of BC, respectively, in Spanish and Chinese people. Rs2228526 and rs2228528 in ERCC6 increase BC susceptibility in Belarussians [181] with the latter also having the same effect in the Taiwanese population [188]. Rs4150667 in GTF2H1 is associated with an increased risk for BC in the Caucasian race [173], rs1805335 in RAD23B increases the risk in the Spanish population [176] and rs2228000 [189–193] and rs2228001 [155, 187, 193–195] in XPC are positively associated with BC in all study populations.
HR is another mechanism that helps maintain the genome’s integrity by repairing double-strand breaks. Therefore, HR deficiencies that result from gene mutations make individuals more susceptible to cancer [196]. Rs861539 in XRCC3 is associated with an increased risk for BC in different populations [160, 197–199]. Rs1799794 and rs861530 in the same gene also increase BC risk in the Chinese population [200]. Rs11571833 in BRCA2 increases BC risk in those of European descents [201]. Rs1805794 in NBN is associated with a high risk for BC [202] while rs8032440 in FANCI and rs3739177 in PNKP make the American population more susceptible to BC [172].
Poly-ADP-ribosylation is a post-translational modification catalyzed by poly(ADP-ribosyl)ation polymerases (PARPs) as a response to DNA damage [203]. Rs1136410 in PARP1 is associated with a high BC risk in the Spanish population [168], whereas rs3219123 and rs12568297 in PARP1, rs1713413 in PARP2, and rs2862907 in PARP4 increase separately the risk for BC in the American population [172].
Cell Cycle
Genes and their corresponding SNPs related to the cellular cycle were separated into three groups consisting of tumor suppressor genes, inhibitors of apoptosis, and other genes associated with BC.
Tumor suppressor genes work by repairing DNA damage, inhibiting cell division, and, in some cases, triggering apoptosis to stop tumor development. Therefore, inactivation or loss of function resulting from mutations in these genes can lead to cancer [204]. An SNP in intron 3 of the TP53 gene was associated with a decreased risk for BC in the American population [183]. Moreover, rs1042522 in the same gene is associated with an increased risk for BC in Asia [205–207]. However, this same SNP showed a protective effect towards BC in the Indian [208] and Brazilian [209] populations, and rs17878362 has the same effect in the American population [210]. Rs2839698 in H19 shows a protective effect towards BC in the Netherlands [211] whereas rs217727 in H19 [212, 213] and rs760805 in RUNX3 [214] increase BC risk in the Chinese population. Finally, rs2073636 in TSC2 is associated with a high risk for BC in the American population [172] and rs17879961 in CHEK2 with a low risk for BC in European descent [201].
Inhibitors of apoptosis are a family of eight proteins. A mutation activating any of the corresponding genes will become a weapon that will be used by tumors to evade apoptosis [215]. Rs2071214 [216], rs8073069 [216], rs9904341 [216–219], and rs3764383 [219] in BIRC5 increase BC risk, while rs17878467 decreases the risk in Asians [216, 219, 220].
Other genes include Cyclin D1 (CCND1) which regulates cell cycle progression through the activation of cyclin-dependent kinases 4 and 6 (CDK4 and CDK6) that lead to Rb protein phosphorylation, thus inactivating it and allowing the cell to progress past the G1/S checkpoint and continue its replication. Over-expression of CCND1 can lead to cancer [221] and the rs9344 in this gene increases the risk for BC [221–225]. Similarly, Cyclin E1 (CCNE1) binds to CDK2 and activates it, allowing the cell to progress and enter phase S [226]. Rs8102137 in CCNE1 widely increases BC development risk [11, 39, 73, 75, 116]. Moreover, the PI3K–AKT–mTOR is implicated in cell growth, tumorigenesis, and cell invasion [227]. Mitochondrial gene POLG polymorphism rs3087374 increases BC risk in the American population [172]. Rs2294008 [11, 37, 39, 73, 79, 228–235] and rs2978974 [70] in the prostate stem cell antigen (PSCA), an inhibitor of cell proliferation, increase widely BC risk.
Cell Architecture
Caveolin-1 (CAV1) is an essential membrane protein expressed in multiple cells. It plays a central role in the formation of caveolae, which are small plasma membrane invaginations involved in signaling and transport. The role of CAV1 in carcinogenesis has been proven. However, the mechanism is still unknown [236]. Rs3807987 and rs7804372 in CAV1 increase the risk for BC in the Taiwanese population [237] while rs1049334 widely increases it. CLTA and CLTC, which encode the light and heavy chains of clathrin, are also associated with BC. Indeed, rs10972786 in CLTA and rs7224631 in CLTC increase the risk of BC in the European population [23]. Clathrin-mediated vesicle pathways include the DNM1 and DNM2 genes. Rs13285411 in DNM1 and rs4804528 in DNM2 increase the risk for BC, while rs4804149 in KANK2 has a significant protective effect towards BC in the European Population [23]. Finally, rs12216499 in RSPH3 widely increases the risk for BC [70], while rs907611 in LSP1 increases the risk for BC in the European population [238] but was found not to be associated with BC in Chinese [70].
Rs738141 in Dynein light chain 4 (DNAL4) implicated in cell motricity increases the risk for BC in the European population [23].
Rs16260 in Cadherin-1 (CDH1) contributes to BC susceptibility in the Chinese population [239].
The extracellular degradation process is mediated by the Matrix Metalloprotease (MMP), which plays a regulatory role in multiple pathways such as apoptosis or angiogenesis. Hence, MMPs participate in carcinogenesis [240]. Conflicting results concerning the association of rs1799750 in MMP1 with BC were obtained [241–245]. Rs243865 in MMP2 has shown an increased BC risk in India [246] and in a meta-analysis conducted by [244]. However, another meta-analysis done by L. Tao et al. has shown, for the same SNP, a protective effect towards BC in the Asian population [245]. Contradictory results have also been reported for rs11568818 in MMP7 [242, 245, 247]. Finally, rs28382575 in MMP11 increases BC risk in Taiwan [248].
Metabolic
Metabolism-related genes were found to be associated with BC. These genes were divided into alcohol metabolism genes, solute carrier transporters (SLC), folate metabolism enzymes, water-soluble vitamin metabolism genes, and other various metabolism-related genes.
Alcohol metabolism requires multiple enzymes. Rs12529 in AKR1C3 decreases BC risk in the Turkish population [249] while Rs4680 in COMT has shown contradictory results [19, 250, 251].
SLC transporters are membrane proteins whose role is to supply cells with nutrients, neurotransmitters, hormones, and drugs. They are usually upregulated in cancer [252]. Rs17674580 in SLC14A [39, 253] and rs1058396 [74], rs10775480 [11, 254], rs10853535 [70, 254], rs17674580 [37, 79, 117], and rs7238033 [39, 70, 254] in SLC14A1 increase BC susceptibility in different populations. Rs1385129 in SLC2A1, also known as Glucose transporter 1 (GLUT1), decreases the carcinogenesis risk in the Chinese population [255]. Rs11871756, rs11077654, rs9913017, and rs4969054 in SLC39A11 increase the risk for BC in different populations [256] and rs2306283 in SLCO1B1 increases the risk in the Japanese population [257].
Mitochondrial folate metabolic enzymes are associated with cancer [258]. Hence, rs1667627 in MTHFD2 increases the risk of developing BC in the American population [89]. Other SNPS in genes involved in the folate metabolism are: rs1801131 [259–262] and rs1801133 [260–265] in MTHFR that are widely classified as risk factors for BC, whereas rs1476413 in the same gene decreases the risk of cancer occurrence [266], rs1805087 in MTR increases the risk for BC in the Tunisian population [262, 267] and rs1801394 in MTRR increases the risk for BC only in Saudi Arabia [18].
SNPs in the metabolism of water-soluble vitamin genes that increase BC risk include rs61330082 in NAMPT in the Chinese population [268], rs4652795 in NMNAT2, rs7636269 in NMNAT3, and rs2304191 in NMRK2 in the European population [23].
Other metabolism genes that are associated with BC are reported in Supplementary Table S1.
Others
Finally, unclassified genes that present various actions and are implicated in different pathways were also found to be associated with BC. These genes are reported under the section named “Others” in Supplementary Table S1.
Not Associated SNPs
Many other SNPs in different genes were also studied and found not to modulate BC risk. Among these, rs3892097 in CYP2D6 was not found to be associated with BC in the Indian [62] and Tunisian [269] populations. Moreover, rs1800629 in TNF-α [270, 271] and rs833061 in VEGFA [122] also showed no association with BC in a meta-analysis. Rs4253211 in ERCC6 [272], rs1042489 in BIRC5 [216] and rs4880 in SOD2 [273] were also not associated with BC. Finally, rs11225395, rs35866072, and rs1940475 in MMP8 [274], as well as rs3918242 [243, 244, 275], rs3918241, rs2250889, rs17576, and rs17577 in MMP9 all showed no association with any risk for carcinogenesis [276]. Other genes and SNPs that strictly showed no association with BC risk are reported in Supplementary Table S1 under the section “No association.”
Discussion
Several SNPs associated with BC are restricted to specific populations. A generalization on an SNP’s potential role in predisposing or protecting an individual from BC cannot be done without testing this specific SNP in a sufficient number of individuals from different races and origins. Some SNPs were studied in different populations, which enabled us to correlate them to BC risk. On the other hand, many other SNPs that were exclusively found not to be associated with BC were only studied in one or two populations. Therefore, the scarcity of the gathered data related to these SNPs renders the interpretation of their correlation with BC challenging and requires broader studies for the validation of their contribution to this type of cancer. For SNPs with conflicting results between studies, GWAS-specific repositories could be useful.
On the other hand, the risk conferred by these variants cannot alone explain the development of BC [277]. In fact, several environmental factors were found to play a role in the pathogenesis of the disease. These include: gender, age, smoking habits, alcohol consumption, and potential environmental exposure. It is also worth mentioning that some SNPs were found to be associated with more invasiveness and recurrence. For example, PSCA rs2294008 was associated with a more invasive disease [234].
Individual SNPs effects on modulating BC risk are minimal. Therefore, studies have shifted towards assessing the polygenic risk score (PRS), as it represents a more accurate representation of an individual’s risk of developing BC. PRS aggregates the effects of multiple SNPs to provide a disease risk prediction [278]. A PRS based on 24 independent GWAS markers showed a fourfold increase in BC risk for both smokers and nonsmokers [5].
Finally, the clinical value of such information has yet to be investigated. In fact, recommendations regarding the clinical management of patients based on their genotypes are still lacking [279].
Conclusion
SNPs are genetic variants that are generally population specific [8]. This review shows that several SNPs were associated with BC, depending on the studied cohorts. The generalization of the link between a variant and BC is not always possible, especially in the absence of data related to large cohorts from different ethnicities. For instance, several SNPs associated with BC are private to specific populations. On the other hand, the involvement of many SNPs in BC was ruled out based on studies focusing on one to two cohorts only. The scarcity of related data urges us to gather all the information we can in order to make it accessible to the scientific community. However, one should consider the complexity of the interpretation of these genomic markers [277], especially that, in many cases, the cumulative effect of several SNPs contributes to modulating the risk for BC [280], in addition to epidemiological or environmental factors such as gender, age, smoking habits, and alcohol consumption. Last but not least, the clinical value of such information has yet to be investigated.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/or.2023.10603/full#supplementary-material.
References
1. GCO. 900-World-Fact-Sheets.Pdf (2021). Available From: https://gco.iarc.fr/today/data/factsheets/populations/900-world-fact-sheets.pdf (Accessed January 25, 2021).
2. GCO. 30-Bladder-Fact-Sheet.Pdf (2021). Available From: https://gco.iarc.fr/today/data/factsheets/cancers/30-Bladder-fact-sheet.pdf (Accessed January 25, 2021).
3. Cumberbatch, MGK, Jubber, I, Black, PC, Esperto, F, Figueroa, JD, Kamat, AM, et al. Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur Urol (2018) 74(6):784–95. doi:10.1016/j.eururo.2018.09.001
4. Wu, K, Wang, X, Xie, Z, Liu, Z, and Lu, Y. N-Acetyltransferase 1 Polymorphism and Bladder Cancer Susceptibility: A Meta-Analysis of Epidemiological Studies. J Int Med Res (2013) 41(1):31–7. doi:10.1177/0300060513476988
5. Koutros, S, Kiemeney, LA, Pal Choudhury, P, Milne, RL, Lopez de Maturana, E, Ye, Y, et al. Genome-Wide Association Study of Bladder Cancer Reveals New Biological and Translational Insights. Eur Urol (2023) 84(1):127–37. doi:10.1016/j.eururo.2023.04.020
6. Audenet, F, Attalla, K, and Sfakianos, JP. The Evolution of Bladder Cancer Genomics: What Have We Learned and How Can We Use It? Urol Oncol Semin Original Invest (2018) 36(7):313–20. doi:10.1016/j.urolonc.2018.02.017
7. Dutt, A, and Beroukhim, R. Single Nucleotide Polymorphism Array Analysis of Cancer. Curr Opin Oncol (2007) 19(1):43–9. doi:10.1097/cco.0b013e328011a8c1
8. Bernig, T, and Chanock, SJ. Challenges of SNP Genotyping and Genetic Variation: Its Future Role in Diagnosis and Treatment of Cancer. Expert Rev Mol Diagn (2006) 6(3):319–31. doi:10.1586/14737159.6.3.319
9. Guo, CC, and Czerniak, B. Bladder Cancer in the Genomic Era. Arch Pathol Lab Med (2019) 143(6):695–704. doi:10.5858/arpa.2018-0329-ra
10. Wu, X, Lin, X, Dinney, CP, Gu, J, and Grossman, HB. Genetic Polymorphism in Bladder Cancer. Front Biosci (2007) 12:192–213. doi:10.2741/2058
11. de Maturana, EL, Rava, M, Anumudu, C, Sáez, O, Alonso, D, and Malats, N. Bladder Cancer Genetic Susceptibility. A Systematic Review. Bladder Cancer (2018) 4(2):215–26. doi:10.3233/blc-170159
12. Wogan, GN, Hecht, SS, Felton, JS, Conney, AH, and Loeb, LA. Environmental and Chemical Carcinogenesis. Semin Cancer Biol (2004) 14(6):473–86. doi:10.1016/j.semcancer.2004.06.010
13. Sankhwar, M, and Sankhwar, SN. Variations in CYP Isoforms and Bladder Cancer: A Superfamily Paradigm. Urol Oncol Semin Original Invest (2014) 32(1):28.e33–28.e40. doi:10.1016/j.urolonc.2012.10.005
14. Wang, Y, Kong, CZ, Zhang, Z, Yang, CM, and Li, J. Relationships Between CYP1A1 Genetic Polymorphisms and Bladder Cancer Risk: A Meta-Analysis. DNA Cel Biol (2014) 33(3):171–81. doi:10.1089/dna.2013.2298
15. Feki-Tounsi, M, Khlifi, R, Louati, I, Fourati, M, Mhiri, MN, Hamza-Chaffai, A, et al. Polymorphisms in XRCC1, ERCC2, and ERCC3 DNA Repair Genes, CYP1A1 Xenobiotic Metabolism Gene, and Tobacco Are Associated With Bladder Cancer Susceptibility in Tunisian Population. Environ Sci Pollut Res (2017) 24(28):22476–84. doi:10.1007/s11356-017-9767-x
16. Grando, JPS, Kuasne, H, Losi-Guembarovski, R, Sant’ana Rodrigues, I, Matsuda, HM, Fuganti, PE, et al. Association Between Polymorphisms in the Biometabolism Genes CYP1A1, GSTM1, GSTT1 and GSTP1 in Bladder Cancer. Clin Exp Med (2009) 9(1):21–8. doi:10.1007/s10238-008-0015-z
17. Lu, Y, Zhang, XL, Xie, L, Li, TJ, He, Y, Peng, QL, et al. Lack of Association Between CYP1A1 Polymorphisms and Risk of Bladder Cancer: A Meta-Analysis. Asian Pac J Cancer Prev (2014) 15(9):4071–7. doi:10.7314/apjcp.2014.15.9.4071
18. Elhawary, NA, Nassir, A, Saada, H, Dannoun, A, Qoqandi, O, Alsharif, A, et al. Combined Genetic Biomarkers Confer Susceptibility to Risk of Urothelial Bladder Carcinoma in a Saudi Population. Dis Markers (2017) 2017:1–11. doi:10.1155/2017/1474560
19. Fontana, L, Delort, L, Joumard, L, Rabiau, N, Bosviel, R, Satih, S, et al. Genetic Polymorphisms in CYP1A1, CYP1B1, COMT, GSTP1 and NAT2 Genes and Association With Bladder Cancer Risk in a French Cohort. Anticancer Res (2009) 29(5):1631–5.
20. Sun, WX, Chen, YH, Liu, ZZ, Xie, JJ, Wang, W, Du, YP, et al. Association Between the CYP1A2 Polymorphisms and Risk of Cancer: A Meta-Analysis. Mol Genet Genomics (2015) 290(2):709–25. doi:10.1007/s00438-014-0956-8
21. Zeng, Y, Jiang, HY, Wei, L, Xu, WD, Wang, YJ, Wang, YD, et al. Association Between the CYP1A2 Rs762551 Polymorphism and Bladder Cancer Susceptibility: A Meta-Analysis Based on Case-Control Studies. Asian Pac J Cancer Prev (2015) 16(16):7249–54. doi:10.7314/apjcp.2015.16.16.7249
22. Vukovic, V, Ianuale, C, Leoncini, E, Pastorino, R, Gualano, MR, Amore, R, et al. Lack of Association Between Polymorphisms in the CYP1A2 Gene and Risk of Cancer: Evidence From Meta-Analyses. BMC Cancer (2016) 16:83. doi:10.1186/s12885-016-2096-5
23. Menashe, I, Figueroa, JD, Garcia-Closas, M, Chatterjee, N, Malats, N, Picornell, A, et al. Large-Scale Pathway-Based Analysis of Bladder Cancer Genome-Wide Association Data From Five Studies of European Background. PLoS One (2012) 7(1):e29396. doi:10.1371/journal.pone.0029396
24. Sankhwar, M, Sankhwar, SN, Abhishek, A, Gupta, N, and Rajender, S. CYP1B1 Gene Polymorphisms Correlate With an Increased Risk of Urinary Bladder Cancer in India. Urol Oncol Semin Original Invest (2016) 34(4):167.e1–167.e8. doi:10.1016/j.urolonc.2015.11.010
25. Salinas-Sánchez, AS, Donate-Moreno, MJ, López-Garrido, MP, Giménez-Bachs, JM, and Escribano, J. Role of CYP1B1 Gene Polymorphisms in Bladder Cancer Susceptibility. J Urol (2012) 187(2):700–6. doi:10.1016/j.juro.2011.10.063
26. Liu, Y, Lin, C, Zhang, A, Song, H, and Fan, C. The CYP1B1 Leu432Val Polymorphism and Risk of Urinary System Cancers. Tumor Biol (2014) 35(5):4719–25. doi:10.1007/s13277-014-1617-6
27. Shi, WX, and Chen, SQ. Frequencies of Poor Metabolizers of Cytochrome P450 2C19 in Esophagus Cancer, Stomach Cancer, Lung Cancer and Bladder Cancer in Chinese Population. World J Gastroenterol (2004) 10(13):1961–3. doi:10.3748/wjg.v10.i13.463
28. Basma, HA, Kobeissi, LH, Jabbour, ME, Moussa, MA, and Dhaini, HR. CYP2E1 and NQO1 Genotypes and Bladder Cancer Risk in a Lebanese Population. Int J Mol Epidemiol Genet (2013) 4(4):207–17.
29. Yin, X, Xiong, W, Wang, Y, Tang, W, Xi, W, Qian, S, et al. Association of CYP2E1 Gene Polymorphisms With Bladder Cancer Risk: A Systematic Review and Meta-Analysis. Medicine (Baltimore) (2018) 97(39):e11910. doi:10.1097/md.0000000000011910
30. Deng, XD, Gao, Q, Zhang, B, Zhang, LX, Zhang, W, Er, ZEM, et al. Functional RsaI/PstI Polymorphism in Cytochrome P450 2E1 Contributes to Bladder Cancer Susceptibility: Evidence From a Meta-Analysis. Asian Pac J Cancer Prev (2014) 15(12):4977–82. doi:10.7314/apjcp.2014.15.12.4977
31. Di Pietro, G, Magno, LAV, and Rios-Santos, F. Glutathione S-Transferases: An Overview in Cancer Research. Expert Opin Drug Metab Toxicol (2010) 6(2):153–70. doi:10.1517/17425250903427980
32. Yu, C, Hequn, C, Longfei, L, Long, W, Zhi, C, Feng, Z, et al. GSTM1 and GSTT1 Polymorphisms Are Associated With Increased Bladder Cancer Risk: Evidence From Updated Meta-Analysis. Oncotarget (2017) 8(2):3246–58. doi:10.18632/oncotarget.13702
33. Yu, Y, Li, X, Liang, C, Tang, J, Qin, Z, Wang, C, et al. The Relationship Between GSTA1, GSTM1, GSTP1, and GSTT1 Genetic Polymorphisms and Bladder Cancer Susceptibility: A Meta-Analysis. Medicine (Baltimore) (2016) 95(37):e4900. doi:10.1097/md.0000000000004900
34. Ouerhani, S, Tebourski, F, Slama, MRB, Marrakchi, R, Rabeh, M, Hassine, LB, et al. The Role of Glutathione Transferases M1 and T1 in Individual Susceptibility to Bladder Cancer in a Tunisian Population. Ann Hum Biol (2006) 33(5–6):529–35. doi:10.1080/03014460600907517
35. Taioli, E, and Raimondi, S. Genetic Susceptibility to Bladder Cancer. The Lancet (2005) 366(9486):610–2. doi:10.1016/s0140-6736(05)67115-2
36. Huang, WW, Chen, DK, Li, LJ, Pan, QW, and Bao, WS. Glutathione S-Transferase M1 and T1 Null Genotypes and Bladder Cancer Risk: A Meta-Analysis in a Single Ethnic Group. J Cancer Res Ther (2018) 14:S993–7. doi:10.4103/0973-1482.191067
37. Wang, M, Chu, H, Lv, Q, Wang, L, Yuan, L, Fu, G, et al. Cumulative Effect of Genome-Wide Association Study-Identified Genetic Variants for Bladder Cancer. Int J Cancer (2014) 135(11):2653–60. doi:10.1002/ijc.28898
38. Kang, HW, Song, PH, Ha, YS, Kim, WT, Kim, YJ, Yun, SJ, et al. Glutathione S-Transferase M1 and T1 Polymorphisms: Susceptibility and Outcomes in Muscle Invasive Bladder Cancer Patients. Eur J Cancer (2013) 49(14):3010–9. doi:10.1016/j.ejca.2013.05.019
39. Dudek, AM, Grotenhuis, AJ, Vermeulen, SH, Kiemeney, LALM, and Verhaegh, GW. Urinary Bladder Cancer Susceptibility Markers. What Do We Know About Functional Mechanisms? Int J Mol Sci (2013) 14(6):12346–66. doi:10.3390/ijms140612346
40. Yuan, JM, Chan, KK, Coetzee, GA, Castelao, JE, Watson, MA, Bell, DA, et al. Genetic Determinants in the Metabolism of Bladder Carcinogens in Relation to Risk of Bladder Cancer. Carcinogenesis (2008) 29(7):1386–93. doi:10.1093/carcin/bgn136
41. Schnakenberg, E, Breuer, R, Werdin, R, Dreikorn, K, and Schloot, W. Susceptibility Genes: GSTM1 and GSTM3 as Genetic Risk Factors in Bladder Cancer. Cytogenet Genome Res (2000) 91(1–4):234–8. doi:10.1159/000056851
42. Törüner, GA, Akyerli, C, Uçar, A, Akı, T, Atsu, N, Ozen, H, et al. Polymorphisms of Glutathione S-Transferase Genes (GSTM1, GSTP1 and GSTT1) and Bladder Cancer Susceptibility in the Turkish Population. Arch Toxicol (2001) 75(8):459–64. doi:10.1007/s002040100268
43. Zhang, R, Xu, G, Chen, W, and Zhang, W. Genetic Polymorphisms of Glutathione S-Transferase M1 and Bladder Cancer Risk: A Meta-Analysis of 26 Studies. Mol Biol Rep (2011) 38(4):2491–7. doi:10.1007/s11033-010-0386-6
44. Zhou, T, Li, HY, Xie, WJ, Zhong, Z, Zhong, H, and Lin, ZJ. Association of Glutathione S-Transferase Gene Polymorphism With Bladder Cancer Susceptibility. BMC Cancer (2018) 18(1):1088. doi:10.1186/s12885-018-5014-1
45. García-Closas, M, Malats, N, Silverman, D, Dosemeci, M, Kogevinas, M, Hein, DW, et al. NAT2 Slow Acetylation, GSTM1 Null Genotype, and Risk of Bladder Cancer: Results From the Spanish Bladder Cancer Study and Meta-Analyses. The Lancet (2005) 366(9486):649–59. doi:10.1016/s0140-6736(05)67137-1
46. Johns, LE, and Houlston, RS. Glutathione S-Transferase Micro1 (GSTM1) Status and Bladder Cancer Risk: A Meta-Analysis. Mutagenesis (2000) 15(5):399–404. doi:10.1093/mutage/15.5.399
47. Lee, SJ, Cho, SH, Park, SK, Kim, SW, Park, MS, Choi, HY, et al. Combined Effect of Glutathione S-Transferase M1 and T1 Genotypes on Bladder Cancer Risk. Cancer Lett (2002) 177(2):173–9. doi:10.1016/s0304-3835(01)00820-5
48. Safarinejad, MR, Safarinejad, S, Shafiei, N, and Safarinejad, S. Association of Genetic Polymorphism of Glutathione S-Transferase (GSTM1, GSTT1, GSTP1) With Bladder Cancer Susceptibility. Urol Oncol Semin Original Invest (2013) 31(7):1193–203. doi:10.1016/j.urolonc.2011.11.027
49. Zhang, R, Xu, G, Chen, W, and Zhang, W. Genetic Polymorphisms of Glutathione S-Transferase P1 and Bladder Cancer Susceptibility in a Chinese Population. Genet Test Mol Biomarkers (2011) 15(1–2):85–8. doi:10.1089/gtmb.2010.0162
50. Song, Y, Chen, J, Liu, K, Zhou, K, Lu, Y, Wang, X, et al. Glutathione S-Transferase Pi 1 (GSTP1) Gene 313 A/G (Rs1695) Polymorphism Is Associated With the Risk of Urinary Bladder Cancer: Evidence From a Systematic Review and Meta-Analysis Based on 34 Case-Control Studies. Gene (2019) 719:144077. doi:10.1016/j.gene.2019.144077
51. Pandith, AA, Lateef, A, Shahnawaz, S, Hussain, A, Malla, TM, Azad, N, et al. GSTP1 Gene Ile105Val Polymorphism Causes an Elevated Risk for Bladder Carcinogenesis in Smokers. Asian Pac J Cancer Prev (2013) 14(11):6375–8. doi:10.7314/apjcp.2013.14.11.6375
52. Wang, Z, Xue, L, Chong, T, Li, H, Chen, H, and Wang, Z. Quantitative Assessment of the Association Between Glutathione S-Transferase P1 Ile105Val Polymorphism and Bladder Cancer Risk. Tumor Biol (2013) 34(3):1651–7. doi:10.1007/s13277-013-0698-y
53. Wu, K, Wang, X, Xie, Z, Liu, Z, and Lu, Y. Glutathione S-Transferase P1 Gene Polymorphism and Bladder Cancer Susceptibility: An Updated Analysis. Mol Biol Rep (2013) 40(1):687–95. doi:10.1007/s11033-012-2109-7
54. Mittal, RD, Srivastava, DSL, Mandhani, A, and Mittal, B. Genetic Polymorphism of Drug Metabolizing Enzymes (CYP2E1, GSTP1) and Susceptibility to Bladder Cancer in North India. Asian Pac J Cancer Prev (2005) 6(1):6–9.
55. Kellen, E, Hemelt, M, Broberg, K, Golka, K, Kristensen, VN, Hung, RJ, et al. Pooled Analysis and Meta-Analysis of the Glutathione S-Transferase P1 Ile 105Val Polymorphism and Bladder Cancer: A HuGE-GSEC Review. Am J Epidemiol (2007) 165(11):1221–30. doi:10.1093/aje/kwm003
56. Djukic, T, Simic, T, Radic, T, Matic, M, Pljesa-Ercegovac, M, Suvakov, S, et al. GSTO1*C/GSTO2*G Haplotype Is Associated With Risk of Transitional Cell Carcinoma of Urinary Bladder. Int Urol Nephrol (2015) 47(4):625–30. doi:10.1007/s11255-015-0933-0
57. Ma, QW, Lin, GF, Chen, JG, and Shen, JH. Polymorphism of Glutathione S-Transferase T1, M1 and P1 Genes in a Shanghai population: Patients With Occupational or Non-Occupational Bladder Cancer. Biomed Environ Sci : BES (2002) 15(3):253–60.
58. Matic, M, Dragicevic, B, Pekmezovic, T, Suvakov, S, Savic-Radojevic, A, Pljesa-Ercegovac, M, et al. Common Polymorphisms in GSTA1, GSTM1 and GSTT1 Are Associated With Susceptibility to Urinary Bladder Cancer in Individuals From Balkan Endemic Nephropathy Areas of Serbia. Tohoku J Exp Med (2016) 240(1):25–30. doi:10.1620/tjem.240.25
59. Goerlitz, D, El Daly, M, Abdel-Hamid, M, Saleh, DA, Goldman, L, El Kafrawy, S, et al. GSTM1, GSTT1 Null Variants, and GPX1 Single Nucleotide Polymorphism Are Not Associated With Bladder Cancer Risk in Egypt. Cancer Epidemiol Biomarkers Prev (2011) 20(7):1552–4. doi:10.1158/1055-9965.epi-10-1306
60. Chen, YC, Xu, L, Guo, YLL, Su, HJJ, Smith, TJ, Ryan, LM, et al. Polymorphisms in GSTT1 and P53 and Urinary Transitional Cell Carcinoma in South-Western Taiwan: A Preliminary Study. Biomarkers (2004) 9(4–5):386–94. doi:10.1080/13547500400010122
61. Gong, M, Dong, W, and An, R. Glutathione S-Transferase T1 Polymorphism Contributes to Bladder Cancer Risk: A Meta-Analysis Involving 50 Studies. DNA Cel Biol (2012) 31(7):1187–97. doi:10.1089/dna.2011.1567
62. Sobti, RC, Al-Badran, AI, Sharma, S, Sharma, SK, Krishan, A, and Mohan, H. Genetic Polymorphisms of CYP2D6, GSTM1, and GSTT1 Genes and Bladder Cancer Risk in North India. Cancer Genet Cytogenet (2005) 156(1):68–73. doi:10.1016/j.cancergencyto.2004.04.001
63. Matic, M, Pekmezovic, T, Djukic, T, Mimic-Oka, J, Dragicevic, D, Krivic, B, et al. GSTA1, GSTM1, GSTP1, and GSTT1 Polymorphisms and Susceptibility to Smoking-Related Bladder Cancer: A Case-Control Study. Urol Oncol Semin Original Invest (2013) 31(7):1184–92. doi:10.1016/j.urolonc.2011.08.005
64. Sanderson, S, Salanti, G, and Higgins, J. Joint Effects of the N-Acetyltransferase 1 and 2 (NAT1 and NAT2) Genes and Smoking on Bladder Carcinogenesis: A Literature-Based Systematic HuGE Review and Evidence Synthesis. Am J Epidemiol (2007) 166(7):741–51. doi:10.1093/aje/kwm167
65. Yassine, IA, Kobeissi, L, Jabbour, ME, and Dhaini, HR. N-Acetyltransferase 1 (NAT1) Genotype: A Risk Factor for Urinary Bladder Cancer in a Lebanese Population. J Oncol (2012) 2012:1–10. doi:10.1155/2012/512976
66. Kobeissi, LH, Yassine, IA, Jabbour, ME, Moussa, MA, and Dhaini, HR. Urinary Bladder Cancer Risk Factors: A Lebanese Case-Control Study. Asian Pac J Cancer Prev (2013) 14(5):3205–11. doi:10.7314/apjcp.2013.14.5.3205
67. Xu, Z, Li, X, Qin, Z, Xue, J, Wang, J, Liu, Z, et al. Association of N-Acetyltransferase 1 Polymorphism and Bladder Cancer Risk: An Updated Meta-Analysis and Trial Sequential Analysis. Int J Biol Markers (2017) 32(3):e297–304. doi:10.5301/ijbm.5000269
68. Hosen, MB, Islam, J, Salam, MA, Islam, MF, Hawlader, MZH, and Kabir, Y. N-Acetyltransferase 2 Gene Polymorphism as a Biomarker for Susceptibility to Bladder Cancer in Bangladeshi Population. Asia Pac J Clin Oncol (2015) 11(1):78–84. doi:10.1111/ajco.12291
69. Ma, QW, Lin, GF, Chen, JG, Xiang, CQ, Guo, WC, Golka, K, et al. Polymorphism of N-Acetyltransferase 2 (NAT2) Gene Polymorphism in Shanghai Population: Occupational and Non-Occupational Bladder Cancer Patient Groups. Biomed Environ Sci : BES (2004) 17(3):291–8.
70. Selinski, S. Urinary Bladder Cancer Risk Variants: Recent Findings and New Challenges of GWAS and Confirmatory Studies. Arch Toxicol (2014) 88(7):1469–75. doi:10.1007/s00204-014-1297-4
71. Izumi, K, Li, Y, Ishiguro, H, Zheng, Y, Yao, JL, Netto, GJ, et al. Expression of UDP-Glucuronosyltransferase 1A in Bladder Cancer: Association With Prognosis and Regulation by Estrogen. Mol Carcinog (2014) 53(4):314–24. doi:10.1002/mc.21978
72. Selinski, S, Lehmann, ML, Blaszkewicz, M, Ovsiannikov, D, Moormann, O, Guballa, C, et al. Rs11892031[A] on Chromosome 2q37 in an Intronic Region of the UGT1A Locus Is Associated With Urinary Bladder Cancer Risk. Arch Toxicol (2012) 86(9):1369–78. doi:10.1007/s00204-012-0854-y
73. Hemminki, K, Bermejo, JL, Ji, J, and Kumar, R. Familial Bladder Cancer and the Related Genes. Curr Opin Urol (2011) 21(5):386–92. doi:10.1097/mou.0b013e32834958ff
74. Zhang, Y, Sun, Y, Chen, T, Hu, H, Xie, W, Qiao, Z, et al. Genetic Variations Rs11892031 and Rs401681 Are Associated With Bladder Cancer Risk in a Chinese Population. Int J Mol Sci (2014) 15(11):19330–41. doi:10.3390/ijms151119330
75. Rothman, N, Garcia-Closas, M, Chatterjee, N, Malats, N, Wu, X, Figueroa, JD, et al. A Multi-Stage Genome-Wide Association Study of Bladder Cancer Identifies Multiple Susceptibility Loci. Nat Genet (2010) 42(11):978–84. doi:10.1038/ng.687
76. Tang, W, Fu, YP, Figueroa, JD, Malats, N, Garcia-Closas, M, Chatterjee, N, et al. Mapping of the UGT1A Locus Identifies an Uncommon Coding Variant That Affects mRNA Expression and Protects From Bladder Cancer. Hum Mol Genet (2012) 21(8):1918–30. doi:10.1093/hmg/ddr619
77. Wang, J, Wu, X, Kamat, A, Barton Grossman, H, Dinney, CP, and Lin, J. Fluid Intake, Genetic Variants of UDP-Glucuronosyltransferases, and Bladder Cancer Risk. Br J Cancer (2013) 108(11):2372–80. doi:10.1038/bjc.2013.190
78. Zhou, Q, Lin, M, Feng, X, Ma, F, Zhu, Y, Liu, X, et al. Targeting CLK3 Inhibits the Progression of Cholangiocarcinoma by Reprogramming Nucleotide Metabolism. J Exp Med (2020) 217(8):e20191779. doi:10.1084/jem.20191779
79. Matsuda, K, Takahashi, A, Middlebrooks, CD, Obara, W, Nasu, Y, Inoue, K, et al. Genome-Wide Association Study Identified SNP on 15q24 Associated With Bladder Cancer Risk in Japanese Population. Hum Mol Genet (2015) 24(4):1177–84. doi:10.1093/hmg/ddu512
80. Chen, DS, and Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity (2013) 39(1):1–10. doi:10.1016/j.immuni.2013.07.012
81. Zou, J, Wu, D, Li, T, Wang, X, Liu, Y, and Tan, S. Association of PD-L1 Gene Rs4143815 C>G Polymorphism and Human Cancer Susceptibility: A Systematic Review and Meta-Analysis. Pathol - Res Pract (2019) 215(2):229–34. doi:10.1016/j.prp.2018.12.002
82. Jaiswal, PK, Singh, V, and Mittal, RD. Cytotoxic T Lymphocyte Antigen 4 (CTLA4) Gene Polymorphism With Bladder Cancer Risk in North Indian Population. Mol Biol Rep (2014) 41(2):799–807. doi:10.1007/s11033-013-2919-2
83. Wang, SS, Hsieh, MJ, Ou, YC, Chen, CS, Li, JR, Hsiao, PC, et al. Impacts of ICAM-1 Gene Polymorphisms on Urothelial Cell Carcinoma Susceptibility and Clinicopathologic Characteristics in Taiwan. Tumor Biol (2014) 35(8):7483–90. doi:10.1007/s13277-014-1934-9
84. Nagarsheth, N, Wicha, MS, and Zou, W. Chemokines in the Cancer Microenvironment and Their Relevance in Cancer Immunotherapy. Nat Rev Immunol (2017) 17(9):559–72. doi:10.1038/nri.2017.49
85. Kucukgergin, C, Isman, FK, Dasdemir, S, Cakmakoglu, B, Sanli, O, Gokkusu, C, et al. The Role of Chemokine and Chemokine Receptor Gene Variants on the Susceptibility and Clinicopathological Characteristics of Bladder Cancer. Gene (2012) 511(1):7–11. doi:10.1016/j.gene.2012.09.011
86. Huang, Y, Chen, H, Wang, J, Bunjhoo, H, Xiong, W, Xu, Y, et al. Relationship Between CCR2-V64i Polymorphism and Cancer Risk: A Meta-Analysis. Gene (2013) 524(1):54–8. doi:10.1016/j.gene.2013.04.011
87. Narter, KF, Agachan, B, Sozen, S, Cincin, ZB, and Isbir, T. CCR2-64I Is a Risk Factor for Development of Bladder Cancer. Genet Mol Res (2010) 9(2):685–92. doi:10.4238/vol9-2gmr829
88. Singh, V, Jaiswal, PK, Kapoor, R, Kapoor, R, and Mittal, RD. Impact of Chemokines CCR5∆32, CXCL12G801A, and CXCR2C1208T on Bladder Cancer Susceptibility in North Indian Population. Tumor Biol (2014) 35(5):4765–72. doi:10.1007/s13277-014-1624-7
89. Andrew, AS, Gui, J, Sanderson, AC, Mason, RA, Morlock, EV, Schned, AR, et al. Bladder Cancer SNP Panel Predicts Susceptibility and Survival. Hum Genet (2009) 125(5–6):527–39. doi:10.1007/s00439-009-0645-6
90. Weng, WC, Huang, YH, Yang, SF, Wang, SS, Kuo, WH, Hsueh, CW, et al. Effect of CD44 Gene Polymorphisms on Risk of Transitional Cell Carcinoma of the Urinary Bladder in Taiwan. Tumor Biol (2016) 37(5):6971–7. doi:10.1007/s13277-015-4566-9
91. Kalvakolanu, DV. The ‘Yin-Yang’ of Cytokines in Cancer. Cytokine (2019) 118:1–2. doi:10.1016/j.cyto.2018.12.013
92. Krajewski, W, Karabon, L, Partyka, A, Tomkiewicz, A, Poletajew, S, Tukiendorf, A, et al. Polymorphisms of Genes Encoding Cytokines Predict the Risk of High-Grade Bladder Cancer and Outcomes of BCG Immunotherapy. Cent Eur J Immunol (2020) 45(1):37–47. doi:10.5114/ceji.2020.94674
93. Jaiswal, PK, Singh, V, Srivastava, P, and Mittal, RD. Association of IL-12, IL-18 Variants and Serum IL-18 With Bladder Cancer Susceptibility in North Indian Population. Gene (2013) 519(1):128–34. doi:10.1016/j.gene.2013.01.025
94. Zhao, T, Wu, X, and Liu, J. Association Between Interleukin-22 Genetic Polymorphisms and Bladder Cancer Risk. Clinics (2015) 70(10):686–90. doi:10.6061/clinics/2015(10)05
95. Galdiero, MR, Marone, G, and Mantovani, A. Cancer Inflammation and Cytokines. Cold Spring Harb Perspect Biol (2018) 10(8):a028662. doi:10.1101/cshperspect.a028662
96. Shi, X, Xie, X, Xun, X, Jia, Y, and Li, S. Associations of IL-10 Genetic Polymorphisms With the Risk of Urologic Cancer: A Meta-Analysis Based on 18,415 Subjects. SpringerPlus (2016) 5(1):2034. doi:10.1186/s40064-016-3705-0
97. Chen, Z, Zhou, W, Dai, M, Wu, Z, and Jin, R. Association Between the Interaction Polymorphisms of Interleukin-10 and Smoking on Patients With Bladder Cancer Risk From a Case-Control Study. Zhonghua Liu Xing Bing Xue Za Zhi Zhonghua Liuxingbingxue Zazhi (2013) 34(2):183–6.
98. Ahirwar, D, Mandhani, A, and Mittal, RD. Interleukin-10 G-1082A and C-819T Polymorphisms as Possible Molecular Markers of Urothelial Bladder Cancer. Arch Med Res (2009) 40(2):97–102. doi:10.1016/j.arcmed.2008.11.006
99. Tang, T, Xue, H, Cui, S, Gong, Z, Fei, Z, Cheng, S, et al. Association of Interleukin-23 Receptor Gene Polymorphisms With Risk of Bladder Cancer in Chinese. Fam Cancer (2014) 13(4):619–23. doi:10.1007/s10689-014-9731-6
100. Ahirwar, DK, Mandhani, A, and Mittal, RD. IL-8 -251 T > A Polymorphism Is Associated With Bladder Cancer Susceptibility and Outcome After BCG Immunotherapy in a Northern Indian Cohort. Arch Med Res (2010) 41(2):97–103. doi:10.1016/j.arcmed.2010.03.005
101. de Maturana, EL, Ye, Y, Calle, ML, Rothman, N, Urrea, V, Kogevinas, M, et al. Application of Multi-SNP Approaches Bayesian LASSO and AUC-RF to Detect Main Effects of Inflammatory-Gene Variants Associated With Bladder Cancer Risk. PLoS One (2013) 8(12):e83745. doi:10.1371/journal.pone.0083745
102. Zhou, B, Zhang, P, Tang, T, Liao, H, Zhang, K, Pu, Y, et al. Polymorphisms and Plasma Levels of IL-27: Impact on Genetic Susceptibility and Clinical Outcome of Bladder Cancer. BMC Cancer (2015) 15:433. doi:10.1186/s12885-015-1459-7
103. Zhang, M, Tan, X, Huang, J, Ke, Z, Ge, Y, Xiong, H, et al. Association of 3 Common Polymorphisms of IL-27 Gene With Susceptibility to Cancer in Chinese: Evidence From an Updated Meta-Analysis of 27 Studies. Med Sci Monitor (2015) 21:2505–13. doi:10.12659/msm.895032
104. Ahirwar, DK, Mandhani, A, Dharaskar, A, Kesarwani, P, and Mittal, RD. Association of Tumour Necrosis Factor-Alpha Gene (T-1031C, C-863A, and C-857T) Polymorphisms With Bladder Cancer Susceptibility and Outcome After Bacille Calmette-Guérin Immunotherapy. BJU Int (2009) 104(6):867–73. doi:10.1111/j.1464-410x.2009.08549.x
105. Gautam, KA, Pooja, S, Sankhwar, SN, Sankhwar, PL, Goel, A, and Rajender, S. c.29C>T Polymorphism in the Transforming Growth Factor-Β1 (TGFB1) Gene Correlates With Increased Risk of Urinary Bladder Cancer. Cytokine (2015) 75(2):344–8. doi:10.1016/j.cyto.2015.05.017
106. Ohadian Moghadam, S, and Nowroozi, MR. Toll-Like Receptors: The Role in Bladder Cancer Development, Progression and Immunotherapy. Scand J Immunol (2019) 90(6):e12818. doi:10.1111/sji.12818
107. Singh, V, Srivastava, N, Kapoor, R, and Mittal, RD. Single-Nucleotide Polymorphisms in Genes Encoding Toll-Like Receptor -2, -3, -4, and -9 in a Case-Control Study With Bladder Cancer Susceptibility in a North Indian Population. Arch Med Res (2013) 44(1):54–61. doi:10.1016/j.arcmed.2012.10.008
108. Shen, Y, Liu, Y, Liu, S, and Zhang, A. Toll-Like Receptor 4 Gene Polymorphisms and Susceptibility to Bladder Cancer. Pathol Oncol Res (2013) 19(2):275–80. doi:10.1007/s12253-012-9579-8
109. Shen, Y, Bu, M, Zhang, A, Liu, Y, and Fu, B. Toll-Like Receptor 4 Gene Polymorphism Downregulates Gene Expression and Involves in Susceptibility to Bladder Cancer. Tumor Biol (2015) 36(4):2779–84. doi:10.1007/s13277-014-2902-0
110. Wan, B, Tan, J, Zeng, Q, He, LY, Gan, Y, Dai, YB, et al. 729G/C Polymorphism in Toll-Like Receptor 4 Results in Increased Susceptibility to Bladder Cancer. Genet Mol Res (2015) 14(4):15482–7. doi:10.4238/2015.november.30.26
111. Cheng, S, Liu, J, Zhang, Y, Lin, Y, Liu, Q, Li, H, et al. Association Detection Between Genetic Variants in the MicroRNA Binding Sites of Toll-Like Receptors Signaling Pathway Genes and Bladder Cancer Susceptibility. Int J Clin Exp Pathol (2014) 7(11):8118–26.
112. Köstner, K, Denzer, N, Müller, CSL, Klein, R, Tilgen, W, and Reichrath, J. The Relevance of Vitamin D Receptor (VDR) Gene Polymorphisms for Cancer: A Review of the Literature. Anticancer Res (2009) 29(9):3511–36.
113. Mittal, RD, Manchanda, PK, Bhat, S, and Bid, HK. Association of Vitamin-D Receptor (Fok-I) Gene Polymorphism With Bladder Cancer in an Indian Population. BJU Int (2007) 99(4):933–7. doi:10.1111/j.1464-410x.2007.06657.x
114. Ben Fradj, MK, Kallel, A, Gargouri, MM, Chehida, MAB, Sallemi, A, Ouanes, Y, et al. Association of FokI Polymorphism of Vitamin D Receptor With Urothelial Bladder Cancer in Tunisians: Role of Tobacco Smoking and Plasma Vitamin D Concentration. Tumor Biol (2016) 37(5):6197–203. doi:10.1007/s13277-015-4496-6
115. Bankhead, A, McMaster, T, Wang, Y, Boonstra, PS, and Palmbos, PL. TP63 Isoform Expression Is Linked With Distinct Clinical Outcomes in Cancer. EBioMedicine (2020) 51:102561. doi:10.1016/j.ebiom.2019.11.022
116. Dudek, AM, Vermeulen, SH, Kolev, D, Grotenhuis, AJ, Kiemeney, LALM, and Verhaegh, GW. Identification of an Enhancer Region Within the TP63/LEPREL1 Locus Containing Genetic Variants Associated With Bladder Cancer Risk. Cell Oncol (2018) 41(5):555–68. doi:10.1007/s13402-018-0393-5
117. Singh, V, Jaiswal, PK, and Mittal, RD. Replicative Study of GWAS TP63C/T, TERTC/T, and SLC14A1C/T With Susceptibility to Bladder Cancer in North Indians1Equal Contribution. Urol Oncol Semin Original Invest (2014) 32(8):1209–14. doi:10.1016/j.urolonc.2014.05.013
118. Wang, M, Wang, M, Zhang, W, Yuan, L, Fu, G, Wei, Q, et al. Common Genetic Variants on 8q24 Contribute to Susceptibility to Bladder Cancer in a Chinese Population. Carcinogenesis (2009) 30(6):991–6. doi:10.1093/carcin/bgp091
119. Wang, M, Du, M, Ma, L, Chu, H, Lv, Q, Ye, D, et al. A Functional Variant in TP63 at 3q28 Associated With Bladder Cancer Risk by Creating an miR-140-5p Binding Site. Int J Cancer (2016) 139(1):65–74. doi:10.1002/ijc.29978
120. Chen, CH, Ho, CH, Kuan-Hua Huang, S, Shen, CH, Wu, CC, and Wang, YH. Association Between VEGF Gene Promoter Polymorphisms and Bladder Cancer: An Updated Meta-Analysis. Cytokine (2020) 131:155112. doi:10.1016/j.cyto.2020.155112
121. Jaiswal, PK, Tripathi, N, Shukla, A, and Mittal, RD. Association of Single Nucleotide Polymorphisms in Vascular Endothelial Growth Factor Gene With Bladder Cancer Risk. Med Oncol (2013) 30(2):509. doi:10.1007/s12032-013-0509-8
122. Song, Y, Yang, Y, Liu, L, and Liu, X. Association Between Five Polymorphisms in Vascular Endothelial Growth Factor Gene and Urinary Bladder Cancer Risk: A Systematic Review and Meta-Analysis Involving 6671 Subjects. Gene (2019) 698:186–97. doi:10.1016/j.gene.2019.02.070
123. Zhang, LF, Ren, KW, Zuo, L, Zou, JG, Song, NH, Mi, YY, et al. VEGF Gene rs3025039C/T and rs833052C/A Variants Are Associated With Bladder Cancer Risk in Asian Descendants. J Cell Biochem (2019) 120(6):10402–12. doi:10.1002/jcb.28324
124. Song, Y, Hu, J, Chen, Q, Guo, J, Zou, Y, Zhang, W, et al. Association Between Vascular Endothelial Growth Factor Rs699947 Polymorphism and the Risk of Three Major Urologic Neoplasms (Bladder Cancer, Prostate Cancer, and Renal Cell Carcinoma): A Meta-Analysis Involving 11,204 Subjects. Gene (2018) 679:241–52. doi:10.1016/j.gene.2018.09.005
125. Ben Wafi, S, Kallel, A, Ben Fradj, MK, Sallemi, A, Ben Rhouma, S, Ben Halima, M, et al. Haplotype-Based Association of Vascular Endothelial Growth Factor Gene Polymorphisms With Urothelial Bladder Cancer Risk in Tunisian Population. J Clin Lab Anal (2018) 32(9):e22610. doi:10.1002/jcla.22610
126. Wei, H, Kamat, AM, Aldousari, S, Ye, Y, Huang, M, Dinney, CP, et al. Genetic Variations in the Transforming Growth Factor Beta Pathway as Predictors of Bladder Cancer Risk. PLoS One (2012) 7(12):e51758. doi:10.1371/journal.pone.0051758
127. Tung, MC, Hsieh, MJ, Wang, SS, Yang, SF, Chen, SS, Wang, SW, et al. Associations of VEGF-C Genetic Polymorphisms With Urothelial Cell Carcinoma Susceptibility Differ Between Smokers and Non-Smokers in Taiwan. PLoS One (2014) 9(3):e91147. doi:10.1371/journal.pone.0091147
128. Lin, A, and Yan, WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol (2018) 9:2164. doi:10.3389/fimmu.2018.02164
129. Castelli, EC, Mendes-Junior, CT, Viana de Camargo, JL, and Donadi, EA. HLA-G Polymorphism and Transitional Cell Carcinoma of the Bladder in a Brazilian Population. Tissue Antigens (2008) 72(2):149–57. doi:10.1111/j.1399-0039.2008.01091.x
130. Chu, H, Wang, M, Jin, H, Lv, Q, Wu, D, Tong, N, et al. EGFR 3’UTR 774T>C Polymorphism Contributes to Bladder Cancer Risk. Mutagenesis (2013) 28(1):49–55. doi:10.1093/mutage/ges051
131. Li, X, Gao, Y, Zhou, H, Xu, W, Li, P, Zhou, J, et al. The Relationship Between Functional Promoter -94 ins/del ATTG Polymorphism in NF-κ B1 Gene and the Risk of Urinary Cancer. Cancer Biomarkers (2016) 16(1):11–7. doi:10.3233/cbm-150536
132. Li, P, Gu, J, Yang, X, Cai, H, Tao, J, Yang, X, et al. Functional Promoter -94 ins/del ATTG Polymorphism in NFKB1 Gene Is Associated With Bladder Cancer Risk in a Chinese Population. PLoS One (2013) 8(8):e71604. doi:10.1371/journal.pone.0071604
133. Tang, T, Cui, S, Deng, X, Gong, Z, Jiang, G, Wang, P, et al. Insertion/Deletion Polymorphism in the Promoter Region of NFKB1 Gene Increases Susceptibility for Superficial Bladder Cancer in Chinese. DNA Cel Biol (2010) 29(1):9–12. doi:10.1089/dna.2009.0937
134. Shen, Y, Lu, Q, Ye, H, Deng, Z, Ma, L, Zhang, Q, et al. Genetic Variant of MAML2 in the NOTCH Signaling Pathway and the Risk of Bladder Cancer: A STROBE-Compliant Study. Medicine (Baltimore) (2020) 99(2):e18725. doi:10.1097/md.0000000000018725
135. Strasser, A, and Vaux, DL. Cell Death in the Origin and Treatment of Cancer. Mol Cel (2020) 78(6):1045–54. doi:10.1016/j.molcel.2020.05.014
136. Verim, L, Timirci-Kahraman, O, Akbulut, H, Akbas, A, Ozturk, T, Turan, S, et al. Functional Genetic Variants in Apoptosis-Associated FAS and FASL Genes and Risk of Bladder Cancer in a Turkish Population. In vivo (Athens, Greece) (2014) 28(3):397–402.
137. Timirci-Kahraman, O, Ozkan, NE, Turan, S, Farooqi, AA, Verim, L, Ozturk, T, et al. Genetic Variants in the Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand and Death Receptor Genes Contribute to Susceptibility to Bladder Cancer. Genet Test Mol Biomarkers (2015) 19(6):309–15. doi:10.1089/gtmb.2015.0050
138. Mittal, RD, Srivastava, P, Mittal, T, Verma, A, Jaiswal, PK, Singh, V, et al. Association of Death Receptor 4, Caspase 3 and 5 Gene Polymorphism With Increased Risk to Bladder Cancer in North Indians. Eur J Surg Oncol (Ejso) (2011) 37(8):727–33. doi:10.1016/j.ejso.2011.05.013
139. Wang, M, Wang, M, Cheng, G, Zhang, Z, Fu, G, and Zhang, Z. Genetic Variants in the Death Receptor 4 Gene Contribute to Susceptibility to Bladder Cancer. Mutat Research/Fundamental Mol Mech Mutagenesis (2009) 661(1–2):85–92. doi:10.1016/j.mrfmmm.2008.11.009
140. Hazra, A, Chamberlain, RM, Grossman, HB, Zhu, Y, Spitz, MR, and Wu, X. Death Receptor 4 and Bladder Cancer Risk. Cancer Res (2003) 63(6):1157–9.
141. Guirado, M, Gil, H, Saenz-Lopez, P, Reinboth, J, Garrido, F, Cozar, JM, et al. Association Between C13ORF31, NOD2, RIPK2 and TLR10 Polymorphisms and Urothelial Bladder Cancer. Hum Immunol (2012) 73(6):668–72. doi:10.1016/j.humimm.2012.03.006
142. Wang, M, Zhang, Z, Tian, Y, Shao, J, and Zhang, Z. A Six-Nucleotide Insertion-Deletion Polymorphism in the CASP8 Promoter Associated With Risk and Progression of Bladder Cancer. Clin Cancer Res (2009) 15(7):2567–72. doi:10.1158/1078-0432.ccr-08-2829
143. Gangwar, R, Mandhani, A, and Mittal, RD. Caspase 9 and Caspase 8 Gene Polymorphisms and Susceptibility to Bladder Cancer in North Indian Population. Ann Surg Oncol (2009) 16(7):2028–34. doi:10.1245/s10434-009-0488-3
144. Hung, SC, Wang, SS, Li, JR, Chen, CS, Yang, CK, Chiu, KY, et al. Effect of HMGB1 Polymorphisms on Urothelial Cell Carcinoma Susceptibility and Clinicopathological Characteristics. Int J Med Sci (2018) 15(14):1731–6. doi:10.7150/ijms.27901
145. Wallace, SS. Base Excision Repair: A Critical Player in Many Games. DNA Repair (2014) 19:14–26. doi:10.1016/j.dnarep.2014.03.030
146. Wang, M, Qin, C, Zhu, J, Yuan, L, Fu, G, Zhang, Z, et al. Genetic Variants of XRCC1, APE1, and ADPRT Genes and Risk of Bladder Cancer. DNA Cel Biol (2010) 29(6):303–11. doi:10.1089/dna.2009.0969
147. Mittal, RD, Mandal, RK, and Gangwar, R. Base Excision Repair Pathway Genes Polymorphism in Prostate and Bladder Cancer Risk in North Indian Population. Mech Ageing Dev (2012) 133(4):127–32. doi:10.1016/j.mad.2011.10.002
148. Mao, Y, Xu, X, Lin, Y, Chen, H, Wu, J, Hu, Z, et al. Quantitative Assessment of the Associations Between XRCC1 Polymorphisms and Bladder Cancer Risk. World J Surg Oncol (2013) 11:58. doi:10.1186/1477-7819-11-58
149. Zhang, F, Wu, JH, Zhao, W, and Liu, HT. XRCC1 Polymorphisms Increase Bladder Cancer Risk in Asians: A Meta-Analysis. Tumor Biol (2013) 34(5):2659–64. doi:10.1007/s13277-013-0816-x
150. Fang, Z, Chen, F, Wang, X, Yi, S, Chen, W, and Ye, G. XRCC1 Arg194Trp and Arg280His Polymorphisms Increase Bladder Cancer Risk in Asian Population: Evidence From a Meta-Analysis. PLoS One (2013) 8(5):e64001. doi:10.1371/journal.pone.0064001
151. Li, S, Peng, Q, Chen, Y, You, J, Chen, Z, Deng, Y, et al. DNA Repair Gene XRCC1 Polymorphisms, Smoking, and Bladder Cancer Risk: A Meta-Analysis. PLoS One (2013) 8(9):e73448. doi:10.1371/journal.pone.0073448
152. Liu, C, Yin, Q, Li, L, Jiao, G, Wang, M, and Wang, Y. XRCC1 Arg194Trp and Arg280His Polymorphisms in Bladder Cancer Susceptibility: A Meta-Analysis. Crit Rev Eukaryot Gene Expr (2013) 23(4):339–54. doi:10.1615/critreveukaryotgeneexpr.2013007781
153. Ricceri, F, Guarrera, S, Sacerdote, C, Polidoro, S, Allione, A, Fontana, D, et al. ERCC1 Haplotypes Modify Bladder Cancer Risk: A Case-Control Study. DNA Repair (2010) 9(2):191–200. doi:10.1016/j.dnarep.2009.12.002
154. Yang, D, Liu, C, Shi, J, Wang, N, Du, X, Yin, Q, et al. Association of XRCC1 Arg399Gln Polymorphism With Bladder Cancer Susceptibility: A Meta-Analysis. Gene (2014) 534(1):17–23. doi:10.1016/j.gene.2013.10.038
155. Fontana, L, Bosviel, R, Delort, L, Guy, L, Chalabi, N, Kwiatkowski, F, et al. DNA Repair Gene ERCC2, XPC, XRCC1, XRCC3 Polymorphisms and Associations With Bladder Cancer Risk in a French Cohort. Anticancer Res (2008) 28(3B):1853–6.
156. Wang, C, Sun, Y, and Han, R. XRCC1 Genetic Polymorphisms and Bladder Cancer Susceptibility: A Meta-Analysis. Urology (2008) 72(4):869–72. doi:10.1016/j.urology.2007.12.059
157. Gao, W, Romkes, M, Zhong, S, Nukui, T, Persad, RA, Smith, PJB, et al. Genetic Polymorphisms in the DNA Repair Genes XPD and XRCC1, P53 Gene Mutations and Bladder Cancer Risk. Oncol Rep (2010) 24(1):257–62. doi:10.3892/or_00000854
158. Zhuo, W, Zhang, L, Cai, L, Zhu, B, and Chen, Z. XRCC1 Arg399Gln Polymorphism and Bladder Cancer Risk: Updated Meta-Analyses Based on 5767 Cases and 6919 Controls. Exp Biol Med (2013) 238(1):66–76. doi:10.1258/ebm.2012.012209
159. Dong, LM, Zhang, XY, Teng, H, Li, MS, and Wang, P. Meta-Analysis Demonstrates No Association Between XRCC1 Arg399Gln Polymorphism and Bladder Cancer Risk. Genet Mol Res (2014) 13(4):9976–85. doi:10.4238/2014.november.28.2
160. Figueroa, JD, Malats, N, Rothman, N, Real, FX, Silverman, D, Kogevinas, M, et al. Evaluation of Genetic Variation in the Double-Strand Break Repair Pathway and Bladder Cancer Risk. Carcinogenesis (2007) 28(8):1788–93. doi:10.1093/carcin/bgm132
161. Mittal, RD, Gangwar, R, Mandal, RK, Srivastava, P, and Ahirwar, DK. Gene Variants of XRCC4 and XRCC3 and Their Association With Risk for Urothelial Bladder Cancer. Mol Biol Rep (2012) 39(2):1667–75. doi:10.1007/s11033-011-0906-z
162. Chang, CH, Chang, CL, Tsai, CW, Wu, HC, Chiu, CF, Wang, RF, et al. Significant Association of an XRCC4 Single Nucleotide Polymorphism With Bladder Cancer Susceptibility in Taiwan. Anticancer Res (2009) 29(5):1777–82.
163. Chang, CH, Chiu, CF, Liang, SY, Wu, HC, Chang, CL, Tsai, CW, et al. Significant Association of Ku80 Single Nucleotide Polymorphisms With Bladder Cancer Susceptibility in Taiwan. Anticancer Res (2009) 29(4):1275–9.
164. Zhi, Y, Yu, J, Liu, Y, Wei, Q, Yuan, F, Zhou, X, et al. Interaction Between Polymorphisms of DNA Repair Genes Significantly Modulated Bladder Cancer Risk. Int J Med Sci (2012) 9(6):498–505. doi:10.7150/ijms.4799
165. Gangwar, R, Ahirwar, D, Mandhani, A, and Mittal, RD. Do DNA Repair Genes OGG1, XRCC3 and XRCC7 Have an Impact on Susceptibility to Bladder Cancer in the North Indian Population? Mutat Research/Genetic Toxicol Environ Mutagenesis (2009) 680(1–2):56–63. doi:10.1016/j.mrgentox.2009.09.008
166. Gangwar, R, Ahirwar, D, Mandhani, A, and Mittal, RD. Influence of XPD and APE1 DNA Repair Gene Polymorphism on Bladder Cancer Susceptibility in North India. Urology (2009) 73(3):675–80. doi:10.1016/j.urology.2008.09.043
167. Liu, C, Yin, Q, Li, L, Zhuang, YZ, Zu, X, and Wang, Y. APE1 Asp148Glu Gene Polymorphism and Bladder Cancer Risk: A Meta-Analysis. Mol Biol Rep (2013) 40(1):171–6. doi:10.1007/s11033-012-2046-5
168. Figueroa, JD, Malats, N, Real, FX, Silverman, D, Kogevinas, M, Chanock, S, et al. Genetic Variation in the Base Excision Repair Pathway and Bladder Cancer Risk. Hum Genet (2007) 121(2):233–42. doi:10.1007/s00439-006-0294-y
169. Ma, L, Chu, H, Wang, M, Shi, D, Zhong, D, Li, P, et al. HOGG1 Ser326Cys Polymorphism Is Associated With Risk of Bladder Cancer in a Chinese Population: A Case-Control Study. Cancer Sci (2012) 103(7):1215–20. doi:10.1111/j.1349-7006.2012.02290.x
170. Ramaniuk, VP, Nikitchenko, NV, Savina, NV, Kuzhir, TD, Rolevich, AI, Krasny, SA, et al. Polymorphism of DNA Repair Genes OGG1, XRCC1, XPD and ERCC6 in Bladder Cancer in Belarus. Biomarkers (2014) 19(6):509–16. doi:10.3109/1354750x.2014.943291
171. Karahalil, B, Kocabas, NA, and Ozçelik, T. DNA Repair Gene Polymorphisms and Bladder Cancer Susceptibility in a Turkish Population. Anticancer Res (2006) 26(6C):4955–8.
172. Xie, H, Gong, Y, Dai, J, Wu, X, and Gu, J. Genetic Variations in Base Excision Repair Pathway and Risk of Bladder Cancer: A Case-Control Study in the United States. Mol Carcinogenesis (2015) 54(1):50–7. doi:10.1002/mc.22073
173. Xing, J, Dinney, CP, Shete, S, Huang, M, Hildebrandt, MA, Chen, Z, et al. Comprehensive Pathway-Based Interrogation of Genetic Variations in the Nucleotide Excision DNA Repair Pathway and Risk of Bladder Cancer. Cancer (2012) 118(1):205–15. doi:10.1002/cncr.26224
174. Jager, M, Blokzijl, F, Kuijk, E, Bertl, J, Vougioukalaki, M, Janssen, R, et al. Deficiency of Nucleotide Excision Repair Is Associated With Mutational Signature Observed in Cancer. Genome Res (2019) 29(7):1067–77. doi:10.1101/gr.246223.118
175. Marteijn, JA, Lans, H, Vermeulen, W, and Hoeijmakers, JHJ. Understanding Nucleotide Excision Repair and Its Roles in Cancer and Ageing. Nat Rev Mol Cel Biol. (2014) 15(7):465–81. doi:10.1038/nrm3822
176. Garcia-Closas, M, Malats, N, Real, FX, Welch, R, Kogevinas, M, Chatterjee, N, et al. Genetic Variation in the Nucleotide Excision Repair Pathway and Bladder Cancer Risk. Cancer Epidemiol Biomarkers Prev (2006) 15(3):536–42. doi:10.1158/1055-9965.epi-05-0749
177. Li, SX, Dai, QS, Chen, SX, Zhang, SD, Liao, XY, Deng, X, et al. Xeroderma Pigmentosum Complementation Group D (XPD) Gene Polymorphisms Contribute to Bladder Cancer Risk: A Meta-Analysis. Tumor Biol (2014) 35(4):3905–15. doi:10.1007/s13277-013-1519-z
178. Wu, Y, and Yang, Y. Complex Association Between ERCC2 Gene Polymorphisms, Gender, Smoking and the Susceptibility to Bladder Cancer: A Meta-Analysis. Tumor Biol (2014) 35(6):5245–57. doi:10.1007/s13277-014-1682-x
179. Shao, J, Gu, M, Xu, Z, Hu, Q, and Qian, L. Polymorphisms of the DNA Gene XPD and Risk of Bladder Cancer in a Southeastern Chinese Population. Cancer Genet Cytogenet (2007) 177(1):30–6. doi:10.1016/j.cancergencyto.2007.05.005
180. Sobti, RC, Kaur, S, Sharma, VL, Singh, SK, Hosseini, SA, and Kler, R. Susceptibility of XPD and RAD51 Genetic Variants to Carcinoma of Urinary Bladder in North Indian Population. DNA Cel Biol (2012) 31(2):199–210. doi:10.1089/dna.2011.1283
181. Savina, NV, Nikitchenko, NV, Kuzhir, TD, Rolevich, AI, Krasny, SA, and Goncharova, RI. The Involvement of ERCC2/XPD and ERCC6/CSB Wild Type Alleles in Protection Against Aging and Cancer. Curr Aging Sci (2018) 11(1):45–54. doi:10.2174/1874609810666170707101548
182. Li, C, Jiang, Z, and Liu, X. XPD Lys(751)Gln and Asp (312)Asn Polymorphisms and Bladder Cancer Risk: A Meta-Analysis. Mol Biol Rep (2010) 37(1):301–9. doi:10.1007/s11033-009-9693-1
183. Wu, X, Gu, J, Grossman, HB, Amos, CI, Etzel, C, Huang, M, et al. Bladder Cancer Predisposition: A Multigenic Approach to DNA-Repair and Cell-Cycle-Control Genes. Am J Hum Genet (2006) 78(3):464–79. doi:10.1086/500848
184. Wang, M, Gu, D, Zhang, Z, Zhou, J, and Zhang, Z. XPD Polymorphisms, Cigarette Smoking, and Bladder Cancer Risk: A Meta-Analysis. J Toxicol Environ Health A (2009) 72(11–12):698–705. doi:10.1080/15287390902841029
185. Li, C, Jiang, Z, and Liu, X. XPD Lys(751)Gln and Asp (312)Asn Polymorphisms and Bladder Cancer Risk: A Meta-Analysis. Mol Biol Rep (2010) 37(1):301–9. doi:10.1007/s11033-009-9693-1
186. Chang, CH, Wang, RF, Tsai, RY, Wu, HC, Wang, CH, Tsai, CW, et al. Significant Association of XPD Codon 312 Single Nucleotide Polymorphism With Bladder Cancer Susceptibility in Taiwan. Anticancer Res (2009) 29(10):3903–7.
187. Wen, H, Ding, Q, Fang, Z, Xia, G, and Fang, J. Population Study of Genetic Polymorphisms and Superficial Bladder Cancer Risk in Han-Chinese Smokers in Shanghai. Int Urol Nephrol (2009) 41(4):855–64. doi:10.1007/s11255-009-9560-y
188. Chang, CH, Chiu, CF, Wang, HC, Wu, HC, Tsai, RY, Tsai, CW, et al. Significant Association of ERCC6 Single Nucleotide Polymorphisms With Bladder Cancer Susceptibility in Taiwan. Anticancer Res (2009) 29(12):5121–4.
189. Zhang, D, Chen, C, Fu, X, Gu, S, Mao, Y, Xie, Y, et al. A Meta-Analysis of DNA Repair Gene XPC Polymorphisms and Cancer Risk. J Hum Genet (2008) 53(1):18–33. doi:10.1007/s10038-007-0215-5
190. Sankhwar, M, Sankhwar, SN, Bansal, SK, Gupta, G, and Rajender, S. Polymorphisms in the XPC Gene Affect Urinary Bladder Cancer Risk: A Case-Control Study, Meta-Analyses and Trial Sequential Analyses. Sci Rep (2016) 6:27018. doi:10.1038/srep27018
191. Qiu, L, Wang, Z, Shi, X, and Wang, Z. Associations Between XPC Polymorphisms and Risk of Cancers: A Meta-Analysis. Eur J Cancer (2008) 44(15):2241–53. doi:10.1016/j.ejca.2008.06.024
192. Wang, Y, Li, Z, Liu, N, and Zhang, G. Association Between CCND1 and XPC Polymorphisms and Bladder Cancer Risk: A Meta-Analysis Based on 15 Case-Control Studies. Tumor Biol (2014) 35(4):3155–65. doi:10.1007/s13277-013-1412-9
193. Dai, QS, Hua, RX, Zeng, RF, Long, JT, and Peng, ZW. XPC Gene Polymorphisms Contribute to Bladder Cancer Susceptibility: A Meta-Analysis. Tumor Biol (2014) 35(1):447–53. doi:10.1007/s13277-013-1062-y
194. Zhang, Y, Wang, X, Zhang, W, and Gong, S. An Association Between XPC Lys939Gln Polymorphism and the Risk of Bladder Cancer: A Meta-Analysis. Tumor Biol (2013) 34(2):973–82. doi:10.1007/s13277-012-0633-7
195. Dou, K, Xu, Q, and Han, X. The Association Between XPC Lys939Gln Gene Polymorphism and Urinary Bladder Cancer Susceptibility: A Systematic Review and Meta-Analysis. Diagn Pathol (2013) 8:112. doi:10.1186/1746-1596-8-112
196. Prakash, R, Zhang, Y, Feng, W, and Jasin, M. Homologous Recombination and Human Health: The Roles of BRCA1, BRCA2, and Associated Proteins. Cold Spring Harb Perspect Biol (2015) 7(4):a016600. doi:10.1101/cshperspect.a016600
197. Ma, Q, Zhao, Y, Wang, S, Zhang, X, Zhang, J, Du, M, et al. Genetic Polymorphisms of XRCC3 Thr241Met (C18067T, Rs861539) and Bladder Cancer Risk: A Meta-Analysis of 18 Research Studies. Tumor Biol (2014) 35(2):1473–80. doi:10.1007/s13277-013-1203-3
198. He, XF, Wei, W, Li, JL, Shen, XL, Ding, Dp, Wang, SL, et al. Association Between the XRCC3 T241M Polymorphism and Risk of Cancer: Evidence From 157 Case-Control Studies. Gene (2013) 523(1):10–9. doi:10.1016/j.gene.2013.03.071
199. Zhu, X, Zhong, Z, Zhang, X, Zhao, X, Xu, R, Ren, W, et al. DNA Repair Gene XRCC3 T241M Polymorphism and Bladder Cancer Risk in a Chinese Population. Genet Test Mol Biomarkers (2012) 16(6):640–3. doi:10.1089/gtmb.2011.0334
200. Zhu, G, Su, H, Lu, L, Guo, H, Chen, Z, Sun, Z, et al. Association of Nineteen Polymorphisms From Seven DNA Repair Genes and the Risk for Bladder Cancer in Gansu Province of China. Oncotarget (2016) 7(21):31372–83. doi:10.18632/oncotarget.9146
201. Ge, Y, Wang, Y, Shao, W, Jin, J, Du, M, Ma, G, et al. Rare Variants in BRCA2 and CHEK2 Are Associated With the Risk of Urinary Tract Cancers. Sci Rep (2016) 6:33542. doi:10.1038/srep33542
202. Zhang, Y, Huang, YS, Lin, WQ, Zhang, SD, Li, QW, Hu, YZ, et al. NBS1 Glu185Gln Polymorphism and Susceptibility to Urinary System Cancer: A Meta-Analysis. Tumor Biol (2014) 35(11):10723–9. doi:10.1007/s13277-014-2346-6
203. Hou, WH, Chen, SH, and Yu, X. Poly-ADP Ribosylation in DNA Damage Response and Cancer Therapy. Mutat Research/Reviews Mutat Res (2019) 780:82–91. doi:10.1016/j.mrrev.2017.09.004
204. Wang, LH, Wu, CF, Rajasekaran, N, and Shin, YK. Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview. Cell Physiol Biochem (2018) 51(6):2647–93. doi:10.1159/000495956
205. Liu, ZH, and Bao, ED. Quantitative Assessment of the Association Between TP53 Arg72Pro Polymorphism and Bladder Cancer Risk. Mol Biol Rep (2013) 40(3):2389–95. doi:10.1007/s11033-012-2319-z
206. Lin, HY, Huang, CH, Yu, TJ, Wu, WJ, Yang, MC, and Lung, FW. p53 Codon 72 Polymorphism Was Associated With Vulnerability, Progression, But Not Prognosis of Bladder Cancer in a Taiwanese Population: An Implication of Structural Equation Modeling to Manage the Risks of Bladder Cancer. Urol Int (2011) 86(3):355–60. doi:10.1159/000323599
207. Hosen, MB, Salam, MA, Islam, MF, Hossain, A, Hawlader, MZH, and Kabir, Y. Association of TP53 Gene Polymorphisms With Susceptibility of Bladder Cancer in Bangladeshi Population. Tumor Biol (2015) 36(8):6369–74. doi:10.1007/s13277-015-3324-3
208. Srivastava, P, Jaiswal, PK, Singh, V, and Mittal, RD. Role of P53 Gene Polymorphism and Bladder Cancer Predisposition in Northern India. Cancer Biomarkers (2011) 8(1):21–8. doi:10.3233/dma-2011-0816
209. Murgel de Castro Santos, LE, Trindade Guilhen, AC, Alves de Andrade, R, Garcia Sumi, L, and Ward, LS. The Role of TP53 PRO47SER and ARG72PRO Single Nucleotide Polymorphisms in the Susceptibility to Bladder Cancer. Urol Oncol Semin Original Invest (2011) 29(3):291–4. doi:10.1016/j.urolonc.2009.03.026
210. Ye, Y, Yang, H, Grossman, HB, Dinney, C, Wu, X, and Gu, J. Genetic Variants in Cell Cycle Control Pathway Confer Susceptibility to Bladder Cancer. Cancer (2008) 112(11):2467–74. doi:10.1002/cncr.23472
211. Verhaegh, GW, Verkleij, L, Vermeulen, SHHM, den Heijer, M, Witjes, JA, and Kiemeney, LA. Polymorphisms in the H19 Gene and the Risk of Bladder Cancer. Eur Urol (2008) 54(5):1118–26. doi:10.1016/j.eururo.2008.01.060
212. Li, Z, and Niu, Y. Association Between lncRNA H19 (Rs217727, Rs2735971 and Rs3024270) Polymorphisms and the Risk of Bladder Cancer in Chinese Population. Minerva Urologica e Nefrologica (2019) 71(2):161–7. doi:10.23736/s0393-2249.18.03004-7
213. Hua, Q, Lv, X, Gu, X, Chen, Y, Chu, H, Du, M, et al. Genetic Variants in lncRNA H19 Are Associated With the Risk of Bladder Cancer in a Chinese Population. Mutagenesis (2016) 31(5):531–8. doi:10.1093/mutage/gew018
214. Zhang, Z, Wang, S, Wang, M, Tong, N, Fu, G, and Zhang, Z. Genetic Variants in RUNX3 and Risk of Bladder Cancer: A Haplotype-Based Analysis. Carcinogenesis (2008) 29(10):1973–8. doi:10.1093/carcin/bgn183
215. Frazzi, R. BIRC3 and BIRC5: Multi-Faceted Inhibitors in Cancer. Cell Biosci (2021) 11(1):8. doi:10.1186/s13578-020-00521-0
216. Zhu, Y, Li, Y, Zhu, S, Tang, R, Liu, Y, and Li, J. Association of Survivin Polymorphisms With Tumor Susceptibility: A Meta-Analysis. PLoS One (2013) 8(9):e74778. doi:10.1371/journal.pone.0074778
217. Mazoochi, T, Karimian, M, Ehteram, H, and Karimian, A. Survivin c.-31G>C (Rs9904341) Gene Transversion and Urinary System Cancers Risk: A Systematic Review and a Meta-Analysis. Personalized Med (2019) 16(1):67–78. doi:10.2217/pme-2018-0053
218. Kawata, N, Tsuchiya, N, Horikawa, Y, Inoue, T, Tsuruta, H, Maita, S, et al. Two Survivin Polymorphisms Are Cooperatively Associated With Bladder Cancer Susceptibility. Int J Cancer (2011) 129(8):1872–80. doi:10.1002/ijc.25850
219. Jaiswal, PK, Goel, A, Mandhani, A, and Mittal, RD. Functional Polymorphisms in Promoter Survivin Gene and Its Association With Susceptibility to Bladder Cancer in North Indian Cohort. Mol Biol Rep (2012) 39(5):5615–21. doi:10.1007/s11033-011-1366-1
220. Fan, S, Meng, J, Zhang, L, Zhang, X, and Liang, C. CAV1 Polymorphisms Rs1049334, Rs1049337, Rs7804372 Might Be the Potential Risk in Tumorigenicity of Urinary Cancer: A Systematic Review and Meta-Analysis. Pathol - Res Pract (2019) 215(1):151–8. doi:10.1016/j.prp.2018.11.009
221. Cortessis, VK, Siegmund, K, Xue, S, Ross, RK, and Yu, MC. A Case-Control Study of Cyclin D1 CCND1 870A-->G Polymorphism and Bladder Cancer. Carcinogenesis (2003) 24(10):1645–50. doi:10.1093/carcin/bgg128
222. Wang, L, Habuchi, T, Takahashi, T, Mitsumori, K, Kamoto, T, Kakehi, Y, et al. Cyclin D1 Gene Polymorphism Is Associated With an Increased Risk of Urinary Bladder Cancer. Carcinogenesis (2002) 23(2):257–64. doi:10.1093/carcin/23.2.257
223. Li, J, Luo, F, Zhang, H, Li, L, and Xu, Y. The CCND1 G870A Polymorphism and Susceptibility to Bladder Cancer. Tumor Biol (2014) 35(1):171–7. doi:10.1007/s13277-013-1021-7
224. Ito, M, Habuchi, T, Watanabe, J, Higashi, S, Nishiyama, H, Wang, L, et al. Polymorphism Within the Cyclin D1 Gene Is Associated With an Increased Risk of Carcinoma In Situ in Patients With Superficial Bladder Cancer. Urology (2004) 64(1):74–8. doi:10.1016/j.urology.2004.03.001
225. Lin, HH, Ke, HL, Hsiao, KH, Tsai, CW, Wu, WJ, Bau, DT, et al. Potential Role of CCND1 G870A Genotype as a Predictor for Urothelial Carcinoma Susceptibility and Muscle-Invasiveness in Taiwan. Chin J Physiol (2011) 54(3):196–202. doi:10.4077/cjp.2011.amm123
226. Teixeira, LK, Reed, SI, and Cyclin, E. Cyclin E Deregulation and Genomic Instability. Adv Exp Med Biol (2017) 1042:527–47. doi:10.1007/978-981-10-6955-0_22
227. Chen, M, Gu, J, Delclos, GL, Killary, AM, Fan, Z, Hildebrandt, MAT, et al. Genetic Variations of the PI3K-AKT-mTOR Pathway and Clinical Outcome in Muscle Invasive and Metastatic Bladder Cancer Patients. Carcinogenesis (2010) 31(8):1387–91. doi:10.1093/carcin/bgq110
228. Wu, X, Ye, Y, Kiemeney, LA, Sulem, P, Rafnar, T, Matullo, G, et al. Genetic Variation in the Prostate Stem Cell Antigen Gene PSCA Confers Susceptibility to Urinary Bladder Cancer. Nat Genet (2009) 41(9):991–5. doi:10.1038/ng.421
229. Wang, S, Tang, J, Wang, M, Yuan, L, and Zhang, Z. Genetic Variation in PSCA and Bladder Cancer Susceptibility in a Chinese Population. Carcinogenesis (2010) 31(4):621–4. doi:10.1093/carcin/bgp323
230. Fu, YP, Kohaar, I, Rothman, N, Earl, J, Figueroa, JD, Ye, Y, et al. Common Genetic Variants in the PSCA Gene Influence Gene Expression and Bladder Cancer Risk. Proc Natl Acad Sci U S A (2012) 109(13):4974–9. doi:10.1073/pnas.1202189109
231. Wang, P, Ye, D, Guo, J, Liu, F, Jiang, H, Gong, J, et al. Genetic Score of Multiple Risk-Associated Single Nucleotide Polymorphisms Is a Marker for Genetic Susceptibility to Bladder Cancer. Genes Chromosomes Cancer (2014) 53(1):98–105. doi:10.1002/gcc.22121
232. Zhao, Y, Gui, ZL, Liao, S, Gao, F, Ge, YZ, and Jia, RP. Prostate Stem Cell Antigen Rs2294008 (C>T) Polymorphism and Bladder Cancer Risk: A Meta-Analysis Based on Cases and Controls. Genet Mol Res (2014) 13(3):5534–40. doi:10.4238/2014.july.25.7
233. Lee, JH, Song, HR, Kim, HN, Kweon, SS, Yun, YW, Choi, JS, et al. Genetic Variation in PSCA Is Associated With Bladder Cancer Susceptibility in a Korean Population. Asian Pac J Cancer Prev (2014) 15(20):8901–4. doi:10.7314/apjcp.2014.15.20.8901
234. Yang, J, Li, W, Zhang, Z, Shen, J, Zhang, N, Yang, M, et al. PSCArs2294008 T Polymorphism Increases the Risk of Bladder Cancer in Bai, Dai, and Han Ethnicity in China and a Potential Mechanism. Genes Genomics (2018) 40(5):531–41. doi:10.1007/s13258-018-0653-9
235. Deng, S, Ren, ZJ, Jin, T, Yang, B, and Dong, Q. Contribution of Prostate Stem Cell Antigen Variation Rs2294008 to the Risk of Bladder Cancer. Medicine (Baltimore) (2019) 98(16):e15179. doi:10.1097/md.0000000000015179
236. Ketteler, J, and Klein, D. Caveolin-1, Cancer and Therapy Resistance. Int J Cancer (2018) 143(9):2092–104. doi:10.1002/ijc.31369
237. Bau, DT, Chang, CH, Tsai, RY, Wang, HC, Wang, RF, Tsai, CW, et al. Significant Association of Caveolin-1 Genotypes With Bladder Cancer Susceptibility in Taiwan. Chin J Physiol (2011) 54(3):153–60. doi:10.4077/cjp.2011.amm009
238. Figueroa, JD, Ye, Y, Siddiq, A, Garcia-Closas, M, Chatterjee, N, Prokunina-Olsson, L, et al. Genome-Wide Association Study Identifies Multiple Loci Associated With Bladder Cancer Risk. Hum Mol Genet (2014) 23(5):1387–98. doi:10.1093/hmg/ddt519
239. Ma, X, Xu, H, Zheng, T, Li, HZ, Shi, TP, Wang, BJ, et al. DNA Polymorphisms in Exon 1 and Promoter of the CDH1 Gene and Relevant Risk of Transitional Cell Carcinoma of the Urinary Bladder. BJU Int (2008) 102(5):633–6. doi:10.1111/j.1464-410x.2008.07634.x
240. Gobin, E, Bagwell, K, Wagner, J, Mysona, D, Sandirasegarane, S, Smith, N, et al. A Pan-Cancer Perspective of Matrix Metalloproteases (MMP) Gene Expression Profile and Their Diagnostic/Prognostic Potential. BMC Cancer (2019) 19(1):581. doi:10.1186/s12885-019-5768-0
241. Tasci, AI, Tugcu, V, Ozbek, E, Ozbay, B, Simsek, A, and Koksal, V. A Single-Nucleotide Polymorphism in the Matrix Metalloproteinase-1 Promoter Enhances Bladder Cancer Susceptibility. BJU Int (2008) 101(4):503–7. doi:10.1111/j.1464-410x.2007.07315.x
242. Srivastava, P, Gangwar, R, Kapoor, R, and Mittal, RD. Bladder Cancer Risk Associated With Genotypic Polymorphism of the Matrix Metalloproteinase-1 and 7 in North Indian Population. Dis Markers (2010) 29(1):37–46. doi:10.1155/2010/149651
243. Wieczorek, E, Reszka, E, Jablonowski, Z, Jablonska, E, Beata Krol, M, Grzegorczyk, A, et al. Genetic Polymorphisms in Matrix Metalloproteinases (MMPs) and Tissue Inhibitors of MPs (TIMPs), and Bladder Cancer Susceptibility. BJU Int (2013) 112(8):1207–14. doi:10.1111/bju.12230
244. Yan, Y, Liang, H, Li, T, Li, M, Li, R, Qin, X, et al. The MMP-1, MMP-2, and MMP-9 Gene Polymorphisms and Susceptibility to Bladder Cancer: A Meta-Analysis. Tumor Biol (2014) 35(4):3047–52. doi:10.1007/s13277-013-1395-6
245. Tao, L, Li, Z, Lin, L, Lei, Y, Hongyuan, Y, Hongwei, J, et al. MMP1, 2, 3, 7, and 9 Gene Polymorphisms and Urinary Cancer Risk: A Meta-Analysis. Genet Test Mol Biomarkers (2015) 19(10):548–55. doi:10.1089/gtmb.2015.0123
246. Srivastava, P, Kapoor, R, and Mittal, RD. Association of Single Nucleotide Polymorphisms in Promoter of Matrix Metalloproteinase-2, 8 Genes With Bladder Cancer Risk in Northern India. Urol Oncol Semin Original Invest (2013) 31(2):247–54. doi:10.1016/j.urolonc.2011.01.001
247. Liao, CH, Chang, WS, Tsai, CW, Hu, PS, Wu, HC, Hsu, SW, et al. Association of Matrix Metalloproteinase-7 Genotypes With the Risk of Bladder Cancer. In Vivo (2018) 32(5):1045–50. doi:10.21873/invivo.11345
248. Li, CC, Hsieh, MJ, Wang, SS, Hung, SC, Lin, CY, Kuo, CW, et al. Impact of Matrix Metalloproteinases 11 Gene Variants on Urothelial Cell Carcinoma Development and Clinical Characteristics. Int J Environ Res Public Health (2020) 17(2):475. doi:10.3390/ijerph17020475
249. Tiryakioglu, NO, and Tunali, NE. Association of AKR1C3 Polymorphisms With Bladder Cancer. Urol J (2016) 13(2):2615–21.
250. Wolpert, BJ, Amr, S, Saleh, DA, Ezzat, S, Gouda, I, Loay, I, et al. Associations Differ by Sex for Catechol-O-Methyltransferase Genotypes and Bladder Cancer Risk in South Egypt. Urol Oncol Semin Original Invest (2012) 30(6):841–7. doi:10.1016/j.urolonc.2010.09.007
251. Chen, Y, Yu, X, Li, T, Yan, H, and Mo, Z. Significant Association of Catechol-O-Methyltransferase Val158Met Polymorphism With Bladder Cancer Instead of Prostate and Kidney Cancer. Int J Biol Markers (2016) 31(2):e110–117. doi:10.5301/jbm.5000204
252. Zhang, Y, and Wang, J. Targeting Uptake Transporters for Cancer Imaging and Treatment. Acta Pharmaceutica Sinica B (2020) 10(1):79–90. doi:10.1016/j.apsb.2019.12.005
253. Rafnar, T, Vermeulen, SH, Sulem, P, Thorleifsson, G, Aben, KK, Witjes, JA, et al. European Genome-Wide Association Study Identifies SLC14A1 as a New Urinary Bladder Cancer Susceptibility Gene. Hum Mol Genet (2011) 20(21):4268–81. doi:10.1093/hmg/ddr303
254. Garcia-Closas, M, Ye, Y, Rothman, N, Figueroa, JD, Malats, N, Dinney, CP, et al. A Genome-Wide Association Study of Bladder Cancer Identifies a New Susceptibility Locus Within SLC14A1, a Urea Transporter Gene on Chromosome 18q12.3. Hum Mol Genet (2011) 20(21):4282–9. doi:10.1093/hmg/ddr342
255. Xu, C, Yang, X, Wang, Y, Ding, N, Han, R, Sun, Y, et al. An Analysis of the Polymorphisms of the GLUT1 Gene in Urothelial Cell Carcinomas of the Bladder and Its Correlation With P53, Ki67 and GLUT1 Expressions. Cancer Gene Ther (2017) 24(7):297–303. doi:10.1038/cgt.2017.17
256. Wu, L, Chaffee, KG, Parker, AS, Sicotte, H, and Petersen, GM. Zinc Transporter Genes and Urological Cancers: Integrated Analysis Suggests a Role for ZIP11 in Bladder Cancer. Tumor Biol (2015) 36(10):7431–7. doi:10.1007/s13277-015-3459-2
257. Bui, HTT, Fujimoto, N, Kubo, T, Inatomi, H, and Matsumoto, T. SLCO1B1, SLCO2B1, and SLCO1B3 Polymorphisms and Susceptibility to Bladder Cancer Risk. Cancer Invest (2014) 32(6):256–61. doi:10.3109/07357907.2014.907421
258. Miyo, M, Konno, M, Colvin, H, Nishida, N, Koseki, J, Kawamoto, K, et al. The Importance of Mitochondrial Folate Enzymes in Human Colorectal Cancer. Oncol Rep (2017) 37(1):417–25. doi:10.3892/or.2016.5264
259. Safarinejad, MR, Shafiei, N, and Safarinejad, S. Genetic Susceptibility of Methylenetetrahydrofolate Reductase (MTHFR) Gene C677T, A1298C, and G1793A Polymorphisms With Risk for Bladder Transitional Cell Carcinoma in Men. Med Oncol (2011) 28(1):S398–412. doi:10.1007/s12032-010-9723-9
260. Kouidhi, S, Rouissi, K, Khedhiri, S, Ouerhani, S, Cherif, M, and Benammar-Elgaaied, A. MTHFR Gene Polymorphisms and Bladder Cancer Susceptibility: A Meta-Analysis Including Race, Smoking Status and Tumour Stage. Asian Pac J Cancer Prev : APJCP (2011) 12(9):2227–32.
261. You, W, Li, Z, Jing, C, Qian-Wei, X, Yu-Ping, Z, Weng-Guang, L, et al. MTHFR C677T and A1298C Polymorphisms Were Associated With Bladder Cancer Risk and Disease Progression: A Meta-Analysis. DNA Cel Biol (2013) 32(5):260–7. doi:10.1089/dna.2012.1931
262. Ouerhani, S, Oliveira, E, Marrakchi, R, Ben Slama, MR, Sfaxi, M, Ayed, M, et al. Methylenetetrahydrofolate Reductase and Methionine Synthase Polymorphisms and Risk of Bladder Cancer in a Tunisian Population. Cancer Genet Cytogenet (2007) 176(1):48–53. doi:10.1016/j.cancergencyto.2007.03.007
263. Izmirli, M, Inandiklioglu, N, Abat, D, Alptekin, D, Demirhan, O, Tansug, Z, et al. MTHFR Gene Polymorphisms in Bladder Cancer in the Turkish Population. Asian Pac J Cancer Prev : APJCP (2011) 12(7):1833–5.
264. Cai, DW, Liu, XF, Bu, RG, Chen, XN, Ning, L, Cheng, Y, et al. Genetic Polymorphisms of MTHFR and Aberrant Promoter Hypermethylation of the RASSF1A Gene in Bladder Cancer Risk in a Chinese Population. J Int Med Res (2009) 37(6):1882–9. doi:10.1177/147323000903700625
265. Lin, J, Spitz, MR, Wang, Y, Schabath, MB, Gorlov, IP, Hernandez, LM, et al. Polymorphisms of Folate Metabolic Genes and Susceptibility to Bladder Cancer: A Case-Control Study. Carcinogenesis (2004) 25(9):1639–47. doi:10.1093/carcin/bgh175
266. Beebe-Dimmer, JL, Iyer, PT, Nriagu, JO, Keele, GR, Mehta, S, Meliker, JR, et al. Genetic Variation in Glutathione S-Transferase Omega-1, Arsenic Methyltransferase and Methylene-Tetrahydrofolate Reductase, Arsenic Exposure and Bladder Cancer: A Case-Control Study. Environ Health (2012) 11:43. doi:10.1186/1476-069x-11-43
267. Rouissi, K, Ouerhani, S, Oliveira, E, Marrakchi, R, Cherni, L, Othman, FB, et al. Polymorphisms in One-Carbon Metabolism Pathway Genes and Risk for Bladder Cancer in a Tunisian Population. Cancer Genet Cytogenet (2009) 195(1):43–53. doi:10.1016/j.cancergencyto.2009.06.007
268. Zhang, K, Zhou, B, Zhang, P, Zhang, Z, Chen, P, Pu, Y, et al. Genetic Variants in NAMPT Predict Bladder Cancer Risk and Prognosis in Individuals From Southwest Chinese Han Group. Tumor Biol (2014) 35(5):4031–40. doi:10.1007/s13277-013-1527-z
269. Ouerhani, S, Marrakchi, R, Bouhaha, R, Ben Slama, MR, Sfaxi, M, Ayed, M, et al. The Role of CYP2D6*4 Variant in Bladder Cancer Susceptibility in Tunisian Patients. Bull Cancer (Paris) (2008) 95(2):E1–4. doi:10.1684/bdc.2008.0583
270. Yang, Z, Lv, Y, Lv, Y, and Wang, Y. Meta-Analysis Shows Strong Positive Association of the TNF-α Gene With Tumor Stage in Bladder Cancer. Urol Int (2012) 89(3):337–41. doi:10.1159/000341701
271. Cai, J, Yang, MY, Hou, N, and Li, X. Association of Tumor Necrosis Factor-α 308G/A Polymorphism With Urogenital Cancer Risk: A Systematic Review and Meta-Analysis. Genet Mol Res (2015) 14(4):16102–12. doi:10.4238/2015.december.7.22
272. Hong, Z, Wu, J, Li, Q, Zhang, S, and Shi, Z. Meta-Analysis Reveals No Significant Association Between ERCC6 Polymorphisms and Bladder Cancer Risk. Int J Biol Markers (2017) 32(1):e113–7. doi:10.5301/jbm.5000236
273. Cao, M, Mu, X, Jiang, C, Yang, G, Chen, H, and Xue, W. Single-Nucleotide Polymorphisms of GPX1 and MnSOD and Susceptibility to Bladder Cancer: A Systematic Review and Meta-Analysis. Tumor Biol (2014) 35(1):759–64. doi:10.1007/s13277-013-1103-6
274. Zhang, LF, Zhu, LJ, Zhang, W, Yuan, W, Song, NH, Zuo, L, et al. MMP-8 C-799 T, Lys460Thr, and Lys87Glu Variants Are Not Related to Risk of Cancer. BMC Med Genet (2019) 20(1):162. doi:10.1186/s12881-019-0890-z
275. Pençe, S, í–zbek, E, Ozan Tiryakioğlu, N, Ersoy Tunali, N, Pençe, HH, and Tunali, H. rs3918242 Variant Genotype Frequency and Increased TIMP-2 and MMP-9 Expression Are Positively Correlated With Cancer Invasion in Urinary Bladder Cancer. Cell Mol Biol (2017) 63(9):46–52. doi:10.14715/cmb/2017.63.9.9
276. Meng, J, Wang, S, Shen, X, Bai, Z, Niu, Q, Ma, D, et al. Polymorphism of MMP-9 Gene Is Not Associated With the Risk of Urinary Cancers: Evidence From an Updated Meta-Analysis. Pathol - Res Pract (2018) 214(12):1966–73. doi:10.1016/j.prp.2018.09.011
277. Mavaddat, N, Pharoah, PDP, Michailidou, K, Tyrer, J, Brook, MN, Bolla, MK, et al. Prediction of Breast Cancer Risk Based on Profiling With Common Genetic Variants. J Natl Cancer Inst (2015) 107(5):djv036. doi:10.1093/jnci/djv036
278. Marees, AT, de Kluiver, H, Stringer, S, Vorspan, F, Curis, E, Marie-Claire, C, et al. A Tutorial on Conducting Genome-Wide Association Studies: Quality Control and Statistical Analysis. Int J Methods Psychiatr Res (2018) 27(2):e1608. doi:10.1002/mpr.1608
279. Tempfer, CB, Hefler, LA, Schneeberger, C, and Huber, JC. How Valid Is Single Nucleotide Polymorphism (SNP) Diagnosis for the Individual Risk Assessment of Breast Cancer? Gynecol Endocrinol (2006) 22(3):155–9. doi:10.1080/09513590600629175
Keywords: bladder cancer, genetic polymorphism, SNP, risk factor, gene
Citation: Kourie HR, Zouein J, Succar B, Mardirossian A, Ahmadieh N, Chouery E, Mehawej C, Jalkh N, kattan J and Nemr E (2023) Genetic Polymorphisms Involved in Bladder Cancer: A Global Review. Oncol. Rev. 17:10603. doi: 10.3389/or.2023.10603
Received: 27 April 2022; Accepted: 06 October 2023;
Published: 06 November 2023.
Edited by:
Carlo Ganini, University of Bari Aldo Moro, ItalyReviewed by:
Hugo Pomares Millan, Dartmouth College, United StatesGaetano Pezzicoli, University of Bari Aldo Moro, Italy
Copyright © 2023 Kourie, Zouein, Succar, Mardirossian, Ahmadieh, Chouery, Mehawej, Jalkh, kattan and Nemr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hampig Raphael Kourie, hampig.kourie@usj.edu.lb
†These authors have contributed equally to this work
 Hampig Raphael Kourie
Hampig Raphael Kourie Joseph Zouein
Joseph Zouein Bahaa Succar
Bahaa Succar Avedis Mardirossian1
Avedis Mardirossian1 Cybel Mehawej
Cybel Mehawej