Abstract
Drought stress poses a serious danger to agricultural production. Recent studies have revealed that most of the chemical methods used in the mitigation of its effects on plant production pose a serious threat to humans and the environment. Therefore, the demand for ecologically friendly solutions to ensure the security of the world’s food supply has increased as a result. Plant growth-promoting rhizobacteria (PGPR) treatment may be advantageous in this situation. Enterobacter mori is a promising rhizobacteria in this regard. However, information on the genome analysis of E. mori linked to the rhizosphere soil of the sorghum plant has not been extensively studied. In this study, we present a genomic lens into functional attributes of E. mori AYS9 isolated from sorghum plants, as well as assess its drought tolerance and plant growth-promoting potentials. Our results showed the drought tolerance and plant growth-promoting potentials of the AYS9. Whole genome sequencing (WGS) results revealed that the genome yielded 4,852,175 bp sequence reads, an average read length of 151 bp, 1,845,357 bp genome size, 67 tRNAs, 3 rRNAs, and a G + C content of 55.5%. The functional genes identified in the genome were linked to processes including phosphate solubilization, iron transport, hormone regulation, nitrogen fixation, and resistance to oxidative and osmotic stress. Also, secondary metabolites supporting bacterial biocontrol properties against phytopathogens, and abiotic stress such as aerobactin-type non-ribosomal peptide siderophore, Stewartan-type ladderane, and Colicin type NRPS were discovered in the AYS9 genome. Our findings however establish that the intricate metabolic pathways mediated by the projected new genes in the bacterial genome may offer a genetic foundation for future understanding of rhizosphere biology and the diverse roles that these genes play in plant development and health.
Introduction
A major abiotic stress that poses a danger to global agricultural productivity is drought (Fadiji et al., 2022b; Fadiji et al., 2022c). Lack of access to fresh water to satisfy the needs of humans and the environment causes and sustains drought, which is a natural phenomenon (Balint et al., 2013; Ojuederie et al., 2019). Since drought is frequently not localised to a certain time or place, monitoring it may be challenging. Lack of water causes serious socioeconomic problems, and crop loss, and is fatal to plants (Salehi-Lisar and Bakhshayeshan-Agdam, 2016; Ahluwalia et al., 2021). Ramakrishna et al. (2019) and Fadiji et al. (2022b) reported that roughly half of the arable land will likely be severely drought-restricted in terms of plant growth by 2052. According to (Mach et al., 2019), drought stress has lowered grain yields by up to 10% over the past 40 years and is expected to have an impact on the output above 50% of most fertile land by the year 2050 (Vinocur and Altman, 2005; Jochum et al., 2019). Research has concentrated on enhancing germplasm and creating techniques for crop management to boost water usage efficiency in order to meet this worldwide challenge in agriculture (Ngumbi and Kloepper, 2016). Due to technological advancements in microbiomics and next-generation sequencing, however, the focus has recently shifted to the use of beneficial microbes that mediate tolerance to drought and increase plant water usage efficiency (Dimkpa et al., 2009; Ngumbi and Kloepper, 2016; Vurukonda et al., 2016).
A sustainable biological strategy to address agricultural production’s water shortage is the use of plant growth-promoting rhizobacteria (PGPR). Intimate and free-living relationships with host plants are quickly established by PGPR in the root rhizosphere. Through a number of processes, these interactions frequently result in an improvement in agricultural yield and the reduction of abiotic and biotic stressors (Vurukonda et al., 2016; Barnawal et al., 2017; Forni et al., 2017). Having been reported as plant disease suppressors, biofertilizers, abiotic stress relievers, and soil toxin removers, PGPR may be extremely important (Naveed et al., 2014; Timmusk et al., 2014). Changes in the structure of the osmoregulation, host root system, management of oxidative stress through the biosynthesis and metabolism of phytohormones or the production of antioxidants for scavenging reactive oxygen species (ROS), the production of large chain exopolysaccharide (EPS), which may act as a humectant, and transcriptional control of the genes responsible for host stress response are some of the mechanisms connected to PGPR-derived drought tolerance (Ngumbi and Kloepper, 2016; Forni et al., 2017; Fadiji et al., 2022a; Fadiji et al., 2022c).
The genus Enterobacter has a total of 23 known species (http://www.bacterio.cict.fr/e/enterobacter.html). Both as pathogenic and beneficial species, Enterobacter species have been implicated in plant diseases and human opportunistic pathogens (Nishijima et al., 2007), as well as essential genetic engineering and plant growth-promoting bacteria (Nie et al., 2002). Others also play vital roles in biocontrol (Zhu et al., 2011). However, only a limited number of Enterobacter species have been identified in plants and soils, although they play numerous functions in plant metabolism and physiology (Zhu et al., 2011; Jochum et al., 2019). For instance, due to their capacity to produce osmoprotective genes and substances, E. mori strains have been widely isolated from the rhizosphere soil of many plants and have been actively involved in the reduction of drought stress in plants. Hence, it may be concluded that E. mori has a potential future in agricultural sustainability.
The identification of key genes in the genome of Enterobacter species might greatly improve these bacteria’s roles in the development of root-bacterial, soil-bacterial, and soil-root interactions for better plant stress response mechanisms (Vurukonda et al., 2018). The lifestyle and operations of the bacteria in the host plants can be impacted by these genes. For instance, the discovery of genes known in the development of biofilm chemotaxis can support plant defense systems and colonization of the root. Based on these functional presumptions, E. mori can be a viable bioagent in the formulation of biostimulants and bioinoculants against drought and for better agricultural yield. In this study, we present a genomic lens into the functional attributes of E. mori AYS9, as well as assess its drought tolerance and plant growth-promoting potentials. As far as we know, this is one of the foremost reports on the WGS of E. mori AYS9 isolated from the rhizosphere of sorghum plants in Southern Africa. Furthermore, this study assessed the plant growth-promoting activities, presence of functional genes and secondary metabolites of the strain.
Materials and Methods
Sampling and Isolation of Rhizobacteria Enterobacter mori AYS9
The rhizobacterium was isolated from 10 g of rhizosphere soil from healthy sorghum plants cultivated in the research farm of the North-West University, South Africa (250 47′25.24056″ S 250 37′30 8.17464″ E). The isolation and characterization of the rhizobacterial isolate were carried out using the method of Sha’arani et al. (2021). A pure rhizobacterium was isolated from 10 g soil tightly adhering to the root of the sorghum through serial dilution and subsequent culturing on Luria–Bertani (LB) agar plates using the method of Majeed et al. (2015). After a series of subculturing, the identified single colonies were characterized using morphological and biochemical tests. The distinct colony was further selected and purified, which was then maintained on a sterile nutrient agar slant for further analyses.
Morphological, Microscopic Appearance, and Biochemical Characterization of Enterobacter mori AYS9
The pure culture of E. mori AYS9 was plated on Nutrient agar and incubated at 28°C for 24 h after which the morphological characteristics of the isolate were examined. The colonies’ morphology, which includes shape, size, pigmentation, and color was properly documented after incubation. Furthermore, the biochemical characterization such as Gram staining, including oxidase, nitrate reduction, Voges Proskauer, catalase, indole, citrate, and starch hydrolysis test by using the standardized method as reported by Clarke and Cowan (1952) and Ahmad et al. (2008). While carbohydrate utilization potentials of the AYS9 were carried out using carbon sources such as galactose, maltose, xylose, lactose, fructose, glucose and sucrose (Sati and Bisht, 2006).
Screening of the Enterobacter mori AYS9 for Drought Tolerance Under Increasing Concentrations of Polyethylene Glycol (PEG6000)
Assessment of the osmotic tolerance of E. mori AYS9 was carried out by documenting their growth within the nutrient-broth medium adjusted with varying PEG6000 concentrations (0%–30%). The cultures were then incubated for 48 h at 28°C under shaking at 200 rpm at the same time (Sandhya et al., 2009). Estimation of the bacterial growth was done by properly recording the cultures’ optical density of the medium at various concentrations of PEG 6000 using Merck’s UV spectrophotometer at 600 nm.
Screening of Enterobacter mori AYS9 for PGP Traits
The standardized methods, as described by Cappuccino and Sherman (1992), as well as Ahmad et al. (2008), were utilized in the testing of E. mori AYS9 for NH3 production in peptone water. The test for phosphate solubilization was done employing a modified procedure as described by Shakeela et al. (2017). The procedures of Ahmad et al. (2008), in addition to those of Masciarelli et al. (2014), were employed to test isolated bacteria for siderophore production. The nutrient broth culture of the isolate was used to inoculate tri-calcium phosphate-supplemented Pikovskaya’s agar. The emergence of yellow coloration around the bacterial inoculation spot, thus, implied a positive result. The nitrogen fixation characteristics of the endophytic bacteria were determined according to Ahmad et al. (2008) in Jensen’s medium. Bacterial growth in the medium indicated the nitrogen-fixing potential.
Furthermore, modified procedures of Loper and Schroth (1986) and Ahmad et al. (2008) were used to determine indole-3-acetic acid (IAA) qualitatively at varying concentrations of tryptophan of 0, 100, and 200 mg within the nutrient broth. The IAA production was signaled by the emergence of pink coloration. Screening of isolate for hydrogen cyanide production (HCN) was also carried out using the procedure described by Ahmad et al. (2008) with slight modifications. The HCN production was indicated by the emergence of orange-to-red coloration. Also, the potential of the isolate to produce 1-aminocyclopropane-1-carboxylate deaminase (ACCd) enzyme was examined on account of the capability to utilize ACC as the only source of nitrogen within the minimal medium (Duan et al., 2009). Bacterial growth on the supplemented ammonium sulphate medium indicated ACCd activity.
Whole-Genome Sequencing (WGS) and Analysis of Enterobacter mori Strain AYS9
The extraction of the DNA of E. mori strain AYS9 was carried out using the Quick-DNA™ Miniprep Kit specific for bacteria and fungi (Zymo Research, United States), using the method stipulated by the manufacturer. DNA quality checks were carried out using a NanoDrop spectrophotometry (Thermo Fischer Scientific, United States). The molecular identification and analysis of the stain AYS9 based on 16S rDNA sequence data was carried out using the method of Majeed et al. (2015). The genome sequencing of the strain AYS9 was performed at the Novogene Company Limited, Singapore.
The WGS of the rhizobacteria strain AYS9 was carried out using the employing of the standard Illumina platform. Furthermore, fragmentation of the genomic DNA of the bacteria was done employing the NEB Ultra II FS kit enzymatic technique. AMPure XP beads were used to assess the resulting fragments of the DNA based on size range (200–650 bp). Also, end-repairing of the DNA was achieved through fragmentation, and the fragment was ligated to Illumina-specifc adapter sequences. Consequently, the sample indexing and selection depending on the size in the second step were carried out. The 4 nM quantity dilution of the standard dilution concentration of the samples was achieved using the fluorometric method. Consequently, sequencing was performed using the pair-end Illumina library and was loaded into the NovaSeq 6000 (2 × 150 bp) instrument for cluster generation and sequencing (300 cycles).
The WGS analysis of the sequence in FASTQ file format was uploaded to the Department of Energy Systems Biology Knowledgebase (KBase) (Arkin et al., 2018). The assessment of the quality of each sequence reads was performed with FastQC tool (version 0.11.5) (Andrews, 2010), while the removal of low-quality bases and sequence adaptor was performed using trimmomatics (version 0.36) (Bolger et al., 2014). Also, sequence reads were assembled by SPAdes (version 3.13.0) (Nurk et al., 2013). Taxonomic annotation was carried out using the GTDB-Tk - v1.7.0 (Chaumeil et al., 2020), while functions annotation was carried out using Rapid Annotations using Subsystems Technology toolkit (RASTtk) (version 1.073) (Aziz et al., 2008) and SEED online server (Overbeek et al., 2014) to group the functions and distribution of the predicted genes. The prediction of functional protein-coding genes was performed using the genomic protein data after processing the genome on the National Center for Biotechnology Information (NCBI) database (Li et al., 2021). All analyses in this study were done using default parameters except otherwise stated. Determination of secondary metabolites in the genome was carried out using antiSMASH (version 6.0.0) (Weber et al., 2015). The circular visualization of the genome with the important genomic feature was plotted using Circular Genome Viewer (CGview) (Arkin et al., 2018), while the phylogeny analysis was performed using MrBayes (Huelsenbeck and Ronquist, 2001). The antiSMASH version 6.0 (Blin et al., 2021) was used for the detection of important metabolites and their biosynthetic gene clusters.
Results
Biochemical, Cultural Features and Drought Tolerance Assay
The cultural, and biochemical characterization alongside plant growth promoting (PGP) features of Enterobacter mori AYS9 from the rhizosphere of the sorghum plant was presented in Table 1. The results showed that AYS9 is Gram-negative, rod-shaped, creamy in colour and catalase positive. Furthermore, strain AYS9 utilizes all the sugars tests carried out and grows optimally between the pH range of 4–10 and temperatures of 20°C–30°C. The plant growth-promoting assay carried out on the strain showed that the strain AYS9 tested positive for all the PGP traits, such as the production of ammonia, phosphate, siderophore, indole-3-acetic acid (IAA), 1-aminocyclopropane-1-carboxylate deaminase (ACCd) as well as nitrogen fixation. But tested negative for hydrogen cyanide production (Table 1). A drought tolerance assay carried out on the strain showed that the strain exhibited increased tolerance to Polyethylene glycol (PEG6000) even to a 30% concentration (Table 2).
TABLE 1
| Cultural and biochemical assay | Plant growth-promoting qualitative tests | ||
|---|---|---|---|
| Characteristics | Result | PGP traits | Results |
| Shape | Rods | Ammonia production | ++ |
| Gram reaction | Gram-negative | Phosphate solubilization | ++ |
| Color | Cream | Siderophore | + |
| Surface texture | Smooth shiny | Nitrogen fixation | + |
| Citrate | + | IAA | + |
| Catalase | + | HCN | − |
| Nitrate | + | ACCd | ++ |
| Maltose | + | ||
| Oxidase | + | ||
| Casein hydrolysis | + | ||
| Fructose | + | ||
| Glucose | + | ||
| Sucrose | + | ||
| Trehalose | + | ||
| Turanose | + | ||
| Galactose | + | ||
| Xylose | + | ||
| Mannitol | + | ||
| Arabinose | + | ||
| Raffinose | + | ||
| Maltose | + | ||
Cultural, biochemical and plant growth-promoting test of Enterobacter mori AYS9.
TABLE 2
| Treatment | O.D at 600 nm |
|---|---|
| 0% | 1.38 ± 0.06 |
| 5% | 1.04 ± 0.01 |
| 10% | 0.88 ± 0.03 |
| 15% | 0.74 ± 0.02 |
| 20% | 0.76 ± 0.01 |
| 25% | 0.89 ± 0.02 |
| 30% | 0.96 ± 0.03 |
Response of Enterobacter mori AYS9 to drought stress amended with various concentrations of PEG-6000.
Values are represented as the mean of 3 replicates (n = 3) ±SE.
WGS Information of the Rhizobacterium Enterobacter mori AYS9
The WGS assessment of E. mori AYS9 showed 7,301,524 bp sequence read count, genome size is 4,852,175 bp, and a G + C content of 55.5% as seen in Table 3. The read length average was 151 bp, while L50 and N50 were 2 and 632,440 bp, respectively while the number of contigs is 34. Furthermore, the genome analysis showcased 4,546 coding sequences, 67 tRNA and 3 rRNA. The circular genome visualization of E. mori strain AYS9 is shown in Figure 1. The statistics of the subsystem revealed 27 subsystem features of the coding protein into functional groups based on the annotated genome grouping by the SEED subsystem in RAST online server. The functional groups of 4,546 protein-coding genes (PCG) were assessed using the RAST and KEGG databases. The 1,732 genes annotated using SEED were classified into cellular components, molecular functions, and biological processes. The six functional groups were found to be prominent these groups are stress response (95 genes), Protein Metabolism (220 genes), carbohydrates (329 genes), Amino Acids and Derivatives (350 genes), respiration (99 genes), cofactor vitamins, prosthetic groups, and pigments (157 genes) (Figure 2).
TABLE 3
| Treatment | Enterobacter mori stain AYS9 |
|---|---|
| Domain | Bacteria |
| Taxonomy | Proteobacteria, Gammaproteobacteria; Enterobacterales, Enterobacteriaceae, Enterobacter |
| Size(bp) | 4,852,175 |
| G + C content (%) | 55.5 |
| N50 | 632,440 |
| L50 | 2 |
| Number of contigs (with PEGs) | 34 |
| Number of subsystems | 372 |
| Number of coding sequences | 4,446 |
| Number of RNAs | 76 |
General characteristics of the genome and annotation details of Enterobacter mori AYS9.
FIGURE 1
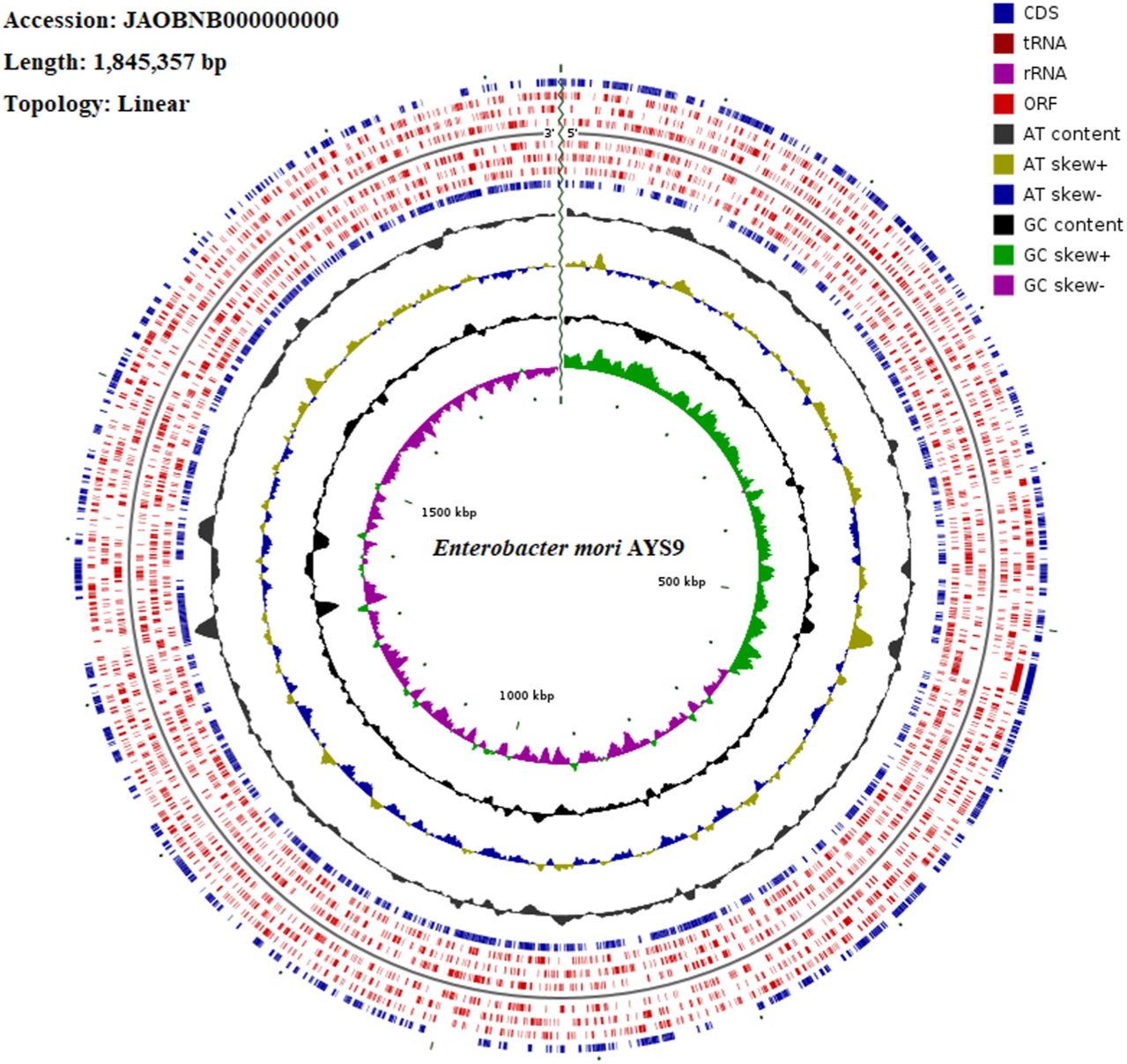
Circular genome visualization of whole-genome Enterobacter mori AYS9.
FIGURE 2
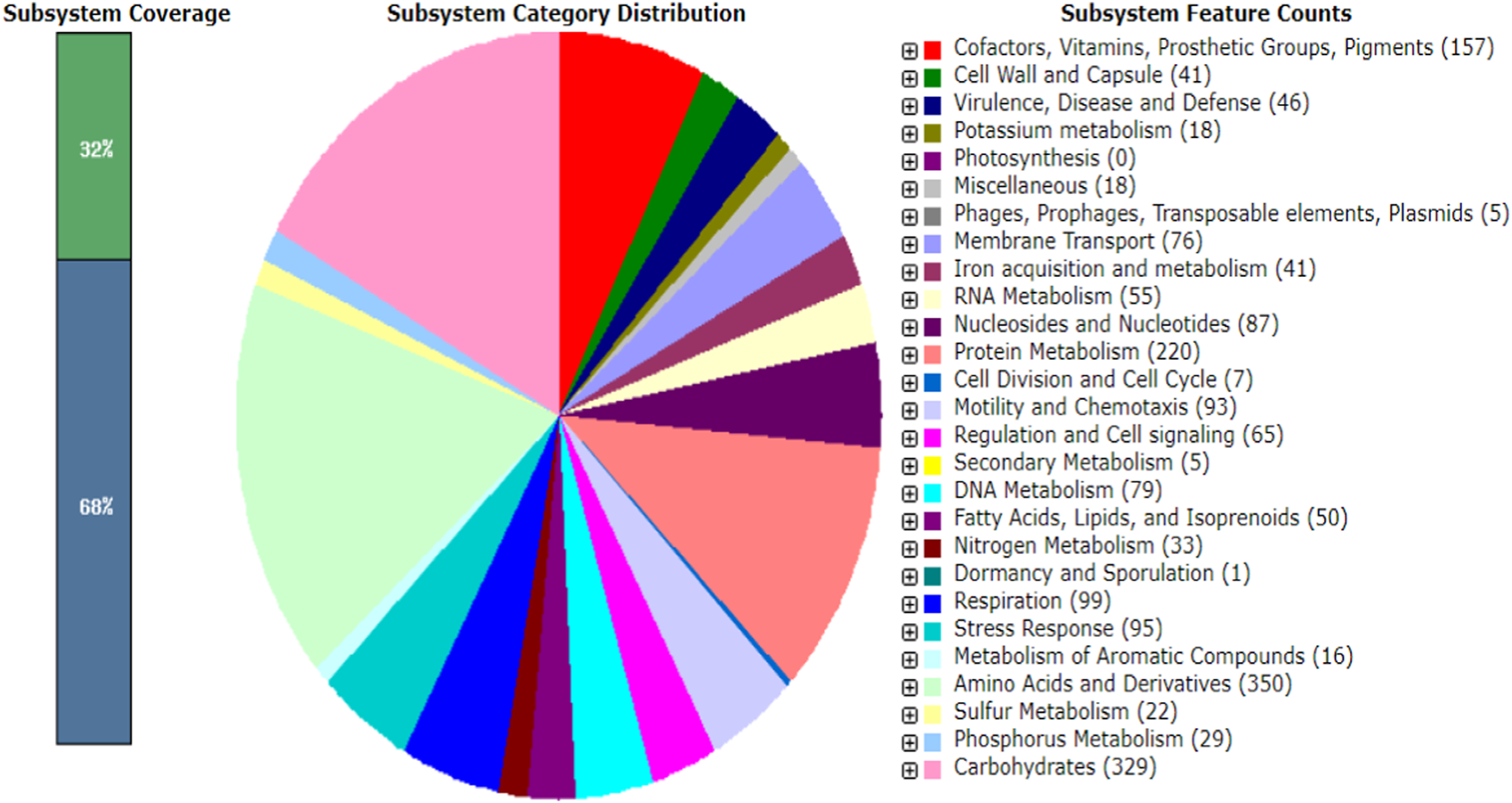
Major PCG of Enterobacter mori strain AYS9 annotated in the RAST and SEED Viewer annotation online server. The green/blue bar on the left shows the coverage of the subsystemin percentage. The blue bar shows the proteins percentage present.
Functional Genes Annotation Information of Enterobacter mori AYS9
Notable metabolic genes were identified in the AYS9 genome (Tables 4–6). Notable genes involved in phosphate solubilization and pathways, such as phosphate assimilation and phosphate transport, are presented in Table 4. The genes, such as fbpC and feoA, coding for iron transport in the Iron (III) transport and Iron (II) transport were detected in strain AYS9 (Table 5). Genes, such as trpA, trpB miaA, miaB speA, speB, speD, speE speG, nirB, nirD, amtB, ureC, ureG, ureF, ureE coding for iron transport, modulation and nitrogen metabolism in the L-tryptophan production, CK biosynthesis, putrescine biosynthesis, spermidine biosynthesis, N1- and N8-acetylspermidine formation, dissimilatory nitrate reduction, ammonia transport, urea degradation and transport respectively are similarly presented in Table 5. Other important genes involved in the mitigation of osmotic and oxidative pathways, such as glycine betaine biosynthesis, proline biosynthesis, glutamate biosynthesis, glycine betaine/proline transport system, glutamate transport, degradation of hydrogen peroxide and degradation of hydrogen peroxide and organic hydroperoxides (Table 6).
TABLE 4
| Genes | Locus Tag | Product | Pathways |
|---|---|---|---|
| ppa | N4Q52_12890 | Inorganic pyrophosphatase | Phosphate assimilation |
| pgl | N4Q52_18515 | 6- phosphogluconolactonase | |
| ppx | N4Q52_08230 | Exopolyphosphatase | |
| pstC | N4Q52_08250 | P (ABC)T permease subunit PstC | |
| phoU | N4Q52_18765 | Phosphate signaling complex protein PhoU | |
| phoA | N4Q52_15190 | Alkaline phosphatase | |
| pstA | N4Q52_18755 | Phosphate ABC transporter (P (ABC)T) permease PstA | Phosphate transport |
| pstB | N4Q52_18760 | P (ABC)T ATP-binding protein PstB | |
| pstS | N4Q52_18745 | P (ABC)T substrate-binding protein PstS |
Notable genes identified in phosphate transport and solubilization.
TABLE 5
| Genes | Locus Tag | Product | Pathways |
|---|---|---|---|
| Iron transport | |||
| FbpC | N4Q52_20740 | Iron (III) transport system ATP-binding protein | Iron (III) transport |
| FeoA | N4Q52_20745 | ferrous iron transport protein A | Iron (II) transport |
| Hormonal modulation | |||
| TrpA | N4Q52_04370 | tryptophan synthase subunit alpha | L-tryptophan production |
| TrpB | N4Q52_04365 | tryptophan synthase subunit beta | |
| MiaA | N4Q52_12590 | tRNA dimethylallyltransferase | CK biosynthesis |
| MiaB | N4Q52_16645 | tRNA-2-methylthio-N6-dimethylallyladenosine synthase | |
| SpeA | N4Q52_10515 | arginine decarboxylase | Putrescine biosynthesis |
| SpeB | N4Q52_10520 | Agmatinase | |
| SpeD | N4Q52_14365 | S-adenosylmethionine decarboxylase | Spermidine biosynthesis |
| SpeE | N4Q52_14370 | Spermidine synthase | |
| SpeG | N4Q52_02295 | Spermidine/Spermine N (1)-acetyltransferase | N1- and N8-acetylspermidine formation |
| Nitrogen metabolism | |||
| NirB | N4Q52_04595 | nitrite reductase (NADH) large subunit | Dissimilatory nitrate reduction |
| NirD | N4Q52_20880 | nitrite reductase (NADH) small subunit | |
| AmtB | N4Q52_15550 | ammonium transporter, Amt family | Ammonia transport |
| UreC | N4Q52_09930 | urease subunit alpha | Urea degradation and transport |
| UreG | N4Q52_09910 | Urease accessory protein UreG | |
| UreF | N4Q52_09915 | Urease accessory protein UreF | |
| UreE | N4Q52_09925 | Urease accessory protein UreE | |
Notable genes identified in iron transport and hormonal modulation.
TABLE 6
| Genes | Locus Tag | Product | Pathways |
|---|---|---|---|
| BetB | N4Q52_16210 | Betaine-aldehyde dehydrogenase | Glycine betaine biosynthesis |
| ProA | N4Q52_16210 | Glutamate-5-semialdehyde dehydrogenase | |
| ProB | N4Q52_15005 | Glutamate 5-kinase | Proline biosynthesis |
| ProC | N4Q52_15215 | Pyrroline-5-carboxylate reductase | |
| GltB | N4Q52_09025 | glutamate synthase (NADPH/NADH) large chain | Glutamate biosynthesis |
| GltD | N4Q52_09020 | glutamate synthase (NADPH/NADH) small chain | |
| GlnA | N4Q52_21360 | Glutamine synthetase | |
| ProX | N4Q52_11795 | Glycine betaine/proline transport system substrate-binding protein | Glycine betaine/proline transport system |
| ProW | N4Q52_11800 | Glycine betaine/proline transport system permease protein | |
| ProV | N4Q52_11805 | Glycine betaine/proline transport system ATP-binding protein | |
| GltP | N4Q52_22150 | proton glutamate symport protein | Glutamate transport |
| KatE | N4Q52_01085 | Catalase | Degradation of hydrogen peroxide |
| Tpx | N4Q52_01085 | Thiol peroxidase | Degradation of hydrogen peroxide and organic hydroperoxides |
| Bcp | N4Q52_08155 | Peroxiredoxin Q/BCP |
Notable genes identified for osmotic and oxidative stress.
Predicted Secondary Metabolite Cluster Genes Through antiSMASH
The secondary metabolite gene clusters identified for strain AYS9 using antiSMASH analysis are reported in Table 7. The identified biosynthetic gene clusters are aerobactin type non-ribosomal peptide siderophore, Stewartan type ladderane, and Colicin type NRPS. The major secondary metabolite gene clusters detected are additional biosynthetic genes, transport-related genes, core biosynthetic genes, regulatory genes, resistance, and other genes (Figures 3A, B). The genes cluster exhibiting 66% and 88% similarity for gene type siderophore and arylpolyene with the most similar known cluster Aerobactin and arylpolygenes node regions 1.1 and 7.1 were also presented (Table 7).
TABLE 7
| Node region | Type | From | To | Most similar known cluster | Similarity | |
|---|---|---|---|---|---|---|
| Rg 1.1 | Siderophore | 1,270,777 | 1,285,205 | Aerobactin | Other | 66% |
| Rg 4.1 | Ladderane | 2009 | 44,567 | Stewartan | Saccharide | 14% |
| Rg 4.2 | NRPS | 322,675 | 366,532 | Colicin | RiPP | 3% |
| Rg 5.1 | Thiopeptide | 130,773 | 157,075 | O-antigen | Saccharide | 14% |
| Rg 7.1 | Arylpolyene | 73,134 | 116,769 | Aryl polygenes | Other | 88% |
Secondary metabolite gene clusters in the genomes of Enterobacter mori strain AYS9.
FIGURE 3
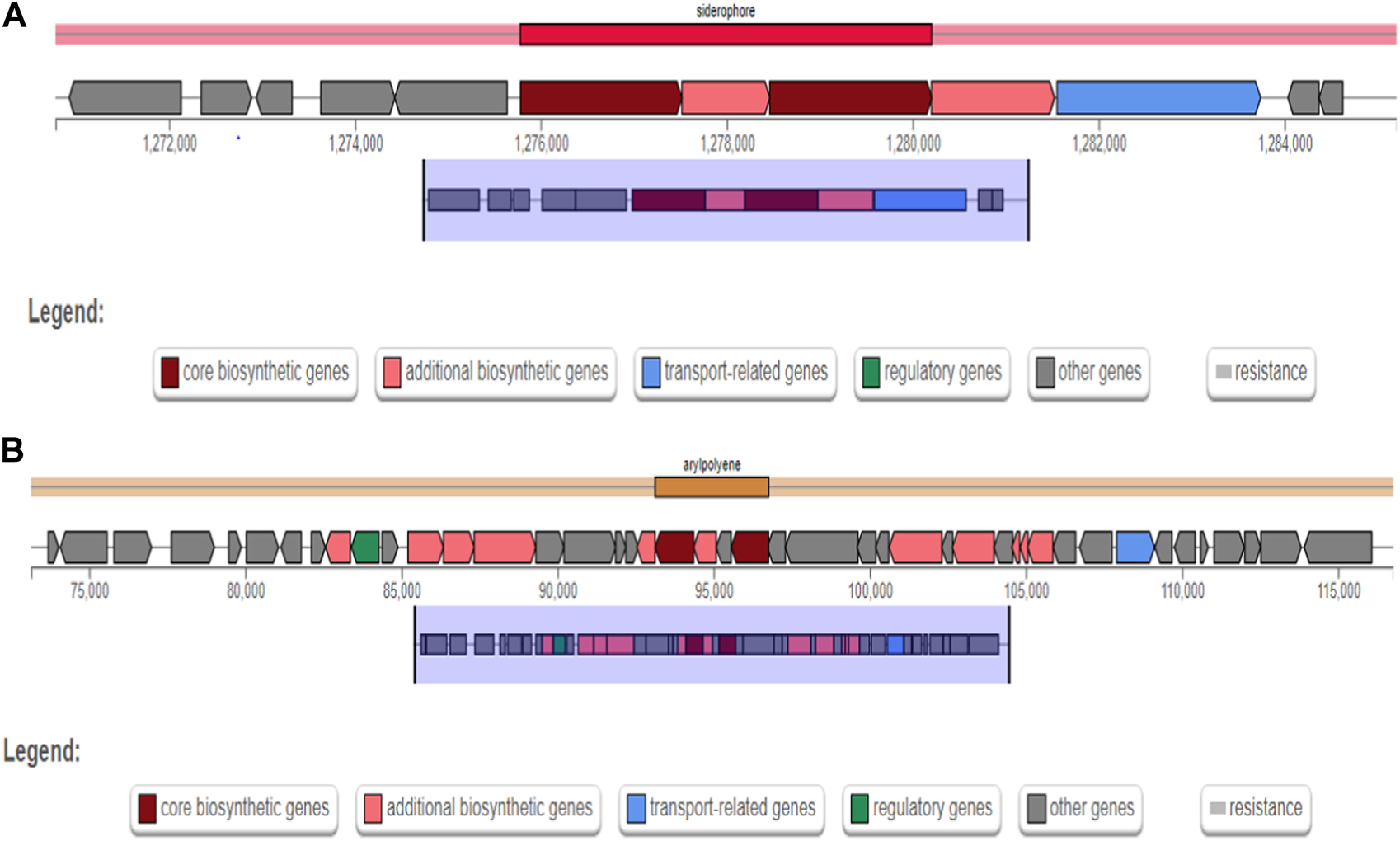
Enterobacter mori strain AYS9 biosynthetic gene clusters for secondary metabolites from antiSmash. (A) siderophore gene clusters and (B) Arylpolene gene clusters.
Discussion
The goal of the study’s design was to identify and describe rhizosphere bacterial strains from the sorghum plant that are capable of stimulating plant development and reducing the effects of drought. A range of hosts, including plants, water, animals, and humans, have been observed to demonstrate an intimate interrelationship with Enterobacter mori, which has been isolated from many ecosystems. In general, it has been found that E. mori, one of the most prevalent species connected to plants, possesses a number of traits that support plant growth (Jha et al., 2011). As a result, knowledge based on their environmental and agricultural value in many crops and plants, such as white mulberry (Zhu et al., 2011), maize (Khan et al., 2022), and peanut (Ludueña et al., 2019) has been reported. This is true to a larger extent due to their potential for plant growth promotion. Whole-genome analysis for identifying several E. mori genes implicated in PGP characteristics, phytohormones, and stress reduction have not yet been investigated. As a result, we presented the first report on the genomic data for E. mori, which was isolated from the sorghum rhizosphere in South Africa.
Enterobacter mori AYS9 was isolated on Luria-Bertani (LB) medium, evaluated on growth-promoting media, and finally sequenced. From both functional and taxonomic data, it was found that the number of genes found in strain AYS9 agrees with those tested in in vitro experiments. In addition to the genes that had been previously assessed, other significant genes of agricultural significance were also found. The strain AYS9 might control plant development in a variety of ways, according to the findings of studies that promoted plant growth. For example, in the solubilization of phosphate, IAA and biosynthesis of siderophore. Our results are consistent with past reports on the properties of Enterobacter strains and other rhizosphere bacteria associated with the root that promote plant development (Kämpfer et al., 2005; Mukherjee and Roy, 2016). Strain AYS9 showed the prospects of being able to mitigate drought stress through its ability to withstand exposure to the PEG 6000. Although its growth was observed to be decreasing initially at 5%–10% concentration of PEG, it started thriving at 20%–30% concentration of PEG. This may be a result of the ability of the rhizobacterium to produce IAA and ACC which has been widely reported to be an important drought-tolerant attribute of rhizobacteria (Fadiji et al., 2022c).
Microbes’ capacity to fix atmospheric nitrogen for the use of the plant is dependent on certain enzymes and genes that they generate (Wang et al., 2004; Peng et al., 2009). Strain AYS9 was found to contain the nitrite reductase enzyme, which codes for by the genes nirB and nirD involved in the dissimilatory nitrate reduction pathway. Other genes discovered are involved in the transport of ammonia (amtB) and urea (ureC, ureG, ureF, and ureE), which may increase the amount of nitrogen in the soil. This outcome is comparable to one from a previous investigation in which flavodoxin and the ammonia transport gene (amtB) were found in MSR2 genome (Nascimento et al., 2020a).
The potential of strain AYS9 to dissolve phosphate under in vitro conditions on tricalcium media was verified by the discovery of many phosphate solubilization and transport-related genes. In a variety of bacterial phyla, gluconic acid (GA) is recognized as a precursor that promotes the solubilization of phosphate. Microbial activities and uses have been influenced by the production of GA by the enzyme glucose-1-dehydrogenase and its nonprotein chemical component pyrroloquinolone quinine (pqq) (Ramachandran et al., 2006). Despite the absence of pqq genes in AYS9 strain, investigations have revealed that the diazotroph bacterial endophyte Herbaspirillum seropedicae Z67 connected to commercial plants expresses heterologous pqq genes (pqqABCDEF) that exhibit phosphate solubilization capacity (Wagh et al., 2014). Similar to this, Klebsiella sp. D5A’s genome was also found to be devoid of pqq genes (Guerrieri et al., 2021). Due to their high affinity and the presence of phosphate transport genes pstABCS, found in strain AYS9 may be better able to absorb phosphates. These results support those made earlier by Guerrieri et al. (2021) and Shariati J et al. (2017) regarding the phosphate transport genes pstABCS expression, on the WG analysis of Klebsiella variicola UC4115 and Pantoea agglomerans P5 respectively. Phosphorus absorption by soil and plant bioavailability may both be improved by the presence of pst genes. The discovery of the phoA gene, which codes for the alkaline phosphatase enzyme involved in numerous phosphorus metabolic processes including phosphate assimilation, was also present in strain AYS9, which was also consistent with the findings for Burkholderia multivorans WS-FJ9 genome (Liu et al., 2020). In addition, the discovery of the genes ppx, pgl and pgl in the AYS9 genome, which respectively encodes for 6-phosphogluconolactonase and exoployphosphatase for the synthesis of d-gluconate and inorganic pyrophosphatase degradation, may have influenced the bacterial strain’s capacity to solubilize phosphate in the plant’s rhizosphere. Our findings, however, are consistent with those made by (Nascimento et al., 2020b), who found that the genome of Bacillus megaterium STB1 contains phoAD genes that boost the bacterial strain’s capacity to solubilize phosphate.
The expression of genes active in the synthesis of IAA and siderophore was found in the rhizosphere bacterial strain AYS9, which is consistent with earlier studies on the PGP traits of rhizobacteria (Peng et al., 2009; Tariq et al., 2014). It has been hypothesized that many pathways, including tryptamine, indole-3-acetonitrile (IAN), indole-3-acetamide, and indole-3-pyruvate (IPA), are involved in the production of IAA in plants and microbes (Spaepen and Vanderleyden, 2011). IAA, a key auxin found in plants, may influence gene expression and biosynthesis in microbes via signaling. Therefore, the production of auxin signal molecules by the microbes may be related to plant responses against naturally occurring harmful pathogens (Spaepen and Vanderleyden, 2011; Fadiji et al., 2022c). In addition, the identical functions of the ipdC gene in Enterobacter cloacae have been discovered (Koga et al., 1991) but were not observed in the genome of AYS9.
It’s interesting to note that strain AYS9 has tryptophan synthase, which is irreversibly implicated in the pyridoxal phosphate pathway and reversibly catalyzes the condensation of serine and indole to create glyceraldehyde-3-phosphate from indole-3-glycerol phosphate (Ireland et al., 2008). The production of IAA by bacteria through the IAN route may be aided by the presence of amidase enzymes. Furthermore, strain AYS9’s participation in two distinct IAA metabolic pathways demonstrated their capacity to produce IAA, which is essential for the creation and growth of plant roots. The AYS9 genome was then found to harbour miaAB genes involved in cytokinin production and transformation. The bacterial genome contains enzyme genes for tRNA dimethylallyltransferases that may improve the generation of iPR—N6-(dimethylallyl) adenosine.
In the Pantoea phytobeneficialis MSR2 genome, various cytokinin genes, such as miaA and miaB, are expressed. These genes change Ipr to 2-methylthio-N6-(dimethylallyl) adenosine and then to 2-methylthio-cis-ribozeatin (Nascimento et al., 2020a). The immediate effects of this specific gene on plant growth promotion imply that cytokinin production may be crucial for promoting plant health and growth (Wani et al., 2016). Despite this, reports on cytokinin genes from this rhizobacterium strain AYS9 are not yet published, necessitating future comparative studies into agriculturally significant E. mori strain AYS9 to learn more about their unique genes. Plant development may be aided by the identification of siderophore and iron-related genes in the E. mori strain AYS9. These genes may increase the accessibility of plants to the soil’s mineral resources. A crucial function in the mineralization of insoluble iron and its bioavailability for plant use may be played by iron transport genes identified in strain AYS9. Additionally, the bacteria’s ability to produce 2,3-butanediol, induce systemic resistance and increase tolerance of the plant to drought have all been connected to their capacity for biocontrol (Madhaiyan et al., 2010; Samaras et al., 2021).
The pathogen-suppressing abilities of rhizobacteria depend on their capacity to create biocontrol agents. Rhizobacteria have used indirect processes in the creation of metabolic chemicals to control phytopathogens (Santoyo et al., 2016; Orozco-Mosqueda et al., 2021). It is possible that siderophore production, which increases the strain AYS9’s antibiosis activity against plant pathogens, is the cause of the strain’s capacity for biocontrol and stress reduction (Maheshwari et al., 2019). It has been determined that the siderophore catecholate present in the bacterial genome is crucial for bacterial adhesion to receptor surfaces, transport and iron chelation (Pedraza et al., 2010). In strain AYS9, iron transport-related genes like fbpC and feoA were discovered. Contrary to earlier findings, no prominent genes involved in siderophore formation and transport that are necessary for the change of chorismate into enterobactin, such as fepABCDG and entABCDEFGHS (Hubrich et al., 2021), we observed in this present study.
The quantity of intracellular polyamines is controlled by production, breakdown, excretion, and absorption from the environment (Kurihara et al., 2011). L-arginine is converted into putrescine via processes that are catalyzed by the enzyme’s arginine decarboxylase and agmatinase. The speA and speB genes encode these enzymes. The two ornithine-decarboxylation enzymes (OCDs), which are either encoded by speC or speF, may also convert L-ornithine into putrescine (Kurihara et al., 2011; Schneider and Wendisch, 2011). Putrescine and decarboxylated S-adenosylmethionine (SAM) served as the building blocks for the production of spermidine, which needs the enzymes speD and speE and were identified in this AYS9 strain. Although it is unclear how these biosynthetic activities are controlled, the intracellular concentration of spermidine appears to be self-regulated (Shah and Swiatlo, 2008).
Plant sensitivity to heat or cold shock may be improved by the presence of osmotic stress-regulating genes. Similarly, to this, the protein-coding genes that control the effects of heat and cold stimuli can work through several gene families and differentiating regulations (Wani et al., 2016). The strain AYS9 included genes such as betB, a component of the glycine betaine biosynthesis pathway known for its osmoprotective properties. This supports past research on Bacillus substilis (Boch et al., 1996; Hussain Wani et al., 2013). Also, genes such as proB and proC involved in proline biosynthesis were identified in the strain AYS9. Additionally, strain AYS9 included genes related to proline biosynthesis, including proB and proC, which is one of the notable mechanisms used by beneficial microbes for the mitigation of drought stress. Pyroline-5-carboxylate synthetase (P5CS1), a stress-induced gene in Arabidopsis, limits L-proline incorporation in chloroplasts (Strizhov et al., 1997; Stein et al., 2011; Fadiji et al., 2021).
In any living creature, glutamine and glutamate act as the primary amino group donors for all nitrogen-containing substances, such as other amino acids and the building blocks for the creation of RNA and DNA. The strain AYS9 has genes for the production and transport of glutamate, including gltB, gltD, and gltP. Glutamate serves as a key intracellular potassium antagonist in addition to its involvement in anabolism (McLaggan et al., 1994). Additionally, certain bacteria and archaea use it as an osmoprotectant (Saum et al., 2006). Our findings are consistent with a previous study on B. subtilis, in which it was shown that glutamate is the precursor to proline, which is present in molar quantities in hyperosmotic circumstances and acts as a compatible solute to protect the cells (Brill et al., 2011).
In this strain AYS9 genome, it was discovered that KatE, which encodes catalase and is involved in the breakdown of hydrogen peroxide, was present. The transgenic indica rice cultivar is more tolerant to salt stress when the catalase genes are overexpressed in E. coli (Moriwaki et al., 2008). Additionally, Escherichia coli strain K12 catalase gene expression increases the jute plant’s resistance to salt stress (Islam et al., 2013). Furthermore, the genome of strain AYS9 had the thiol peroxidase gene, which was shown to degrade organic hydroperoxides and hydrogen peroxide. This gene is well-known for its function in the biocontrol of wheat blast by Bacillus spp. This function is related to induced systemic resistance and the generation of antimicrobial chemicals in host plants like tomatoes (Baier and Dietz, 1996; El-Gaied et al., 2013).
Conclusion
In-depth Information on the prominent plant growth-promoting genes present in E. mori AYS9 with diverse roles and potential to mitigate drought were explored in this study. Some significant regulatory and plant growth-promoting genes were discovered by genome analysis. Notable genes that encourage plant development were found in E. mori. Their importance and promise in agriculture cannot be underestimated. The numerous genes discovered had biotechnological significance in the interactions between microorganisms and plants, in plant resistance to environmental stress (drought, osmotic and oxidative), and in organic compounds that could improve soil health and plant growth and the yield of the crop. The existence of these notable genes has important implications for understanding E. mori AYS9’s genome PGP activities alongside showing promise in establishing sustainable agriculture. Additionally, according to the data on E. mori AYS9, future research is encouraged to take advantage of its potential as a prospective option for creating bioinoculants to deal with abiotic stress-related agricultural issues in the future, most importantly through field trials.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
AF, AA, and OB conceived the ideas. AF collected the data and developed the manuscript. AA and OB provided technical input and proofread the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was funded by the National Research Foundation, South Africa (UID123634 and UID132595) awarded to OB.
Acknowledgments
AF and AA gratefully acknowledged the North-West University for a Postdoctoral fellowship. OB appreciates the research grant (UID132595) that has supported this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Ahluwalia O. Singh P. C. Bhatia R. (2021). A Review on Drought Stress in Plants: Implications, Mitigation and the Role of Plant Growth Promoting Rhizobacteria. Resour. Environ. Sustain.5, 100032. 10.1016/j.resenv.2021.100032
2
Ahmad F. Ahmad I. Khan M. (2008). Screening of Free-Living Rhizospheric Bacteria for Their Multiple Plant Growth Promoting Activities. Microbiol. Res.163, 173–181. 10.1016/j.micres.2006.04.001
3
Andrews S. (2010). “FastQC: a Quality Control Tool for High Throughput Sequence Data,” in Babraham Bioinformatics (Cambridge, United Kingdom: Babraham Institute).
4
Arkin A. P. Cottingham R. W. Henry C. S. Harris N. L. Stevens R. L. Maslov S. et al (2018). KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol.36, 566–569. 10.1038/nbt.4163
5
Aziz R. K. Bartels D. Best A. A. Dejongh M. Disz T. Edwards R. A. et al (2008). The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genomics9, 75–15. 10.1186/1471-2164-9-75
6
Baier M. Dietz K.-J. (1996). Primary Structure and Expression of Plant Homologues of Animal and Fungal Thioredoxin-dependent Peroxide Reductases and Bacterial Alkyl Hydroperoxide Reductases. Plant Mol. Biol.31, 553–564. 10.1007/bf00042228
7
Balint Z. Mutua F. Muchiri P. Omuto C. T. (2013). “Monitoring Drought with the Combined Drought Index in Kenya,” in Developments in Earth Surface Processes. Editors ParonP.OlagoD. O.OmutoC. T. (Amsterdam: Elsevier), 16, 2–374.
8
Barnawal D. Bharti N. Pandey S. S. Pandey A. Chanotiya C. S. Kalra A. (2017). Plant Growth‐promoting Rhizobacteria Enhance Wheat Salt and Drought Stress Tolerance by Altering Endogenous Phytohormone Levels and TaCTR1/TaDREB2 Expression. Physiol. Plant.161, 502–514. 10.1111/ppl.12614
9
Blin K. Shaw S. Kloosterman A. M. Charlop-Powers Z. Van Wezel G. P. Medema M. H. et al (2021). antiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res.49, W29–W35. 10.1093/nar/gkab335
10
Boch J. Kempf B. Schmid R. Bremer E. (1996). Synthesis of the Osmoprotectant glycine Betaine in Bacillus Subtilis: Characterization of the gbsAB Genes. J. Bacteriol.178, 5121–5129. 10.1128/jb.178.17.5121-5129.1996
11
Bolger A. M. Lohse M. Usadel B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics30, 2114–2120. 10.1093/bioinformatics/btu170
12
Brill J. Hoffmann T. Bleisteiner M. Bremer E. (2011). Osmotically Controlled Synthesis of the Compatible Solute Proline Is Critical for Cellular Defense of Bacillus Subtilis against High Osmolarity. J. Bacteriol.193, 5335–5346. 10.1128/jb.05490-11
13
Cappuccino J. Sherman N. (1992). Microbiology: A Laboratory Manual. New York: Pearson Education Limited, 125–179.
14
Chaumeil P.-A. Mussig A. J. Hugenholtz P. Parks D. H. (2020). GTDB-tk: a Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics36 (6), 1925–1927. Oxford University Press. 10.1093/bioinformatics/btz848
15
Clarke P. H. Cowan S. (1952). Biochemical Methods for Bacteriology. Microbiology6, 187–197. 10.1099/00221287-6-1-2-187
16
Dimkpa C. Merten D. Svatoš A. Büchel G. Kothe E. (2009). Siderophores Mediate Reduced and Increased Uptake of Cadmium by Streptomyces Tendae F4 and Sunflower (Helianthus Annuus), Respectively. J. Appl. Microbiol.107, 1687–1696. 10.1111/j.1365-2672.2009.04355.x
17
Duan J. Müller K. M. Charles T. C. Vesely S. Glick B. R. (2009). 1-aminocyclopropane-1-carboxylate (ACC) Deaminase Genes in Rhizobia from Southern Saskatchewan. Microb. Ecol.57, 421–422. 10.1007/s00248-009-9493-0
18
El-Gaied L. F. Abu El-Heba G. A. El-Sherif N. A. (2013). Effect of Growth Hormones on Some Antioxidant Parameters and Gene Expression in Tomato. GM Crops Food4, 67–73. 10.4161/gmcr.24324
19
Fadiji A. E. Ayangbenro A. S. Babalola O. O. (2021). Unveiling the Putative Functional Genes Present in Root-Associated Endophytic Microbiome from Maize Plant Using the Shotgun Approach. J. Appl. Genet.62, 339–351. 10.1007/s13353-021-00611-w
20
Fadiji A. E. Babalola O. O. Santoyo G. Perazzolli M. (2022a). The Potential Role of Microbial Biostimulants in the Amelioration of Climate Change-Associated Abiotic Stresses on Crops. Front. Microbiol.12, 829099. 10.3389/fmicb.2021.829099
21
Fadiji A. E. Orozco-Mosqueda M. D. C. Santos-Villalobos S. D. L. Santoyo G. Babalola O. O. (2022b). Recent Developments in the Application of Plant Growth-Promoting Drought Adaptive Rhizobacteria for Drought Mitigation. Plants11, 3090. 10.3390/plants11223090
22
Fadiji A. E. Santoyo G. Yadav A. N. Babalola O. O. (2022c). Efforts towards Overcoming Drought Stress in Crops: Revisiting the Mechanisms Employed by Plant Growth-Promoting Bacteria. Front. Microbiol.13, 962427. 10.3389/fmicb.2022.962427
23
Forni C. Duca D. Glick B. R. (2017). Mechanisms of Plant Response to Salt and Drought Stress and Their Alteration by Rhizobacteria. Plant Soil410, 335–356. 10.1007/s11104-016-3007-x
24
Guerrieri M. C. Fiorini A. Fanfoni E. Tabaglio V. Cocconcelli P. S. Trevisan M. et al (2021). Integrated Genomic and Greenhouse Assessment of a Novel Plant Growth-Promoting Rhizobacterium for Tomato Plant. Front. Plant Sci.12, 660620. 10.3389/fpls.2021.660620
25
Hubrich F. Müller M. Andexer J. N. (2021). Chorismate-and Isochorismate Converting Enzymes: Versatile Catalysts Acting on an Important Metabolic Node. Chem. Commun.57, 2441–2463. 10.1039/d0cc08078k
26
Huelsenbeck J. P. Ronquist F. (2001). MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics17, 754–755. 10.1093/bioinformatics/17.8.754
27
Hussain Wani S. Brajendra Singh N. Haribhushan A. Iqbal Mir J. (2013). Compatible Solute Engineering in Plants for Abiotic Stress Tolerance-Role of glycine Betaine. Curr. genomics14, 157–165. 10.2174/1389202911314030001
28
Ireland C. Peekhaus N. Lu P. Sangari R. Zhang A. Masurekar P. et al (2008). The Tryptophan Synthetase Gene TRP1 of Nodulisporium sp. Molecular Characterization and its Relation to Nodulisporic Acid A Production. Appl. Microbiol. Biotechnol.79, 451–459. 10.1007/s00253-008-1440-3
29
Islam M. S. Azam M. S. Sharmin S. Sajib A. A. Alam M. Reza M. S. et al (2013). Improved Salt Tolerance of Jute Plants Expressing the katE Gene from Escherichia coli. Turk J. Biol.37, 206–211. 10.3906/biy-1205-52
30
Jha C. K. Aeron A. Patel B. V. Maheshwari D. K. Saraf M. (2011). “Enterobacter: Role in Plant Growth Promotion,” in Bacteria in Agrobiology: Plant Growth Responses. Editor MaheshwariD. (Berlin, Heidelberg: Springer), 159–182. 10.1007/978-3-642-20332-9_8
31
Jochum M. D. Mcwilliams K. L. Borrego E. J. Kolomiets M. V. Niu G. Pierson E. A. et al (2019). Bioprospecting Plant Growth-Promoting Rhizobacteria that Mitigate Drought Stress in Grasses. Front. Microbiol.10, 2106. 10.3389/fmicb.2019.02106
32
Kämpfer P. Ruppel S. Remus R. (2005). Enterobacter Radicincitans Sp. nov., a Plant Growth Promoting Species of the Family Enterobacteriaceae. Syst. Appl. Microbiol.28, 213–221. 10.1016/j.syapm.2004.12.007
33
Khan M. A. Shahzad H. Waheed M. Anwar F. Adil M. Qazi I. et al (2022). Rhizobacterial Inoculation Effect on Yield Contributing Parameters of Maize (Zea Mays). Nat. Volatiles Essent. Oils9, 237–240. 10.1016/j.jgar.2022.06.028
34
Koga J. Adachi T. Hidaka H. (1991). IAA Biosynthetic Pathway from Tryptophan via Indole-3-Pyruvic Acid in Enterobacter cloacae. Agric. Biol. Chem.55, 701–706. 10.1271/bbb1961.55.701
35
Kurihara S. Suzuki H. Oshida M. Benno Y. (2011). A Novel Putrescine Importer Required for Type 1 Pili-Driven Surface Motility Induced by Extracellular Putrescine in Escherichia coli K-12. J. Biol. Chem.286, 10185–10192. 10.1074/jbc.m110.176032
36
Li W. O’neill K. R. Haft D. H. Dicuccio M. Chetvernin V. Badretdin A. et al (2021). RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline Reach with Protein Family Model Curation. Nucleic Acids Res.49, D1020–D1028. 10.1093/nar/gkaa1105
37
Liu Y.-Q. Wang Y.-H. Kong W.-L. Liu W.-H. Xie X.-L. Wu X.-Q. (2020). Identification, Cloning and Expression Patterns of the Genes Related to Phosphate Solubilization in Burkholderia Multivorans WS-FJ9 under Different Soluble Phosphate Levels. Amb. Express10, 108–111. 10.1186/s13568-020-01032-4
38
Loper J. Schroth M. (1986). Influence of Bacterial Sources of Indole-3-Acetic Acid on Root Elongation of Sugar Beet. Phytopathology76, 386–389. 10.1094/phyto-76-386
39
Ludueña L. M. Anzuay M. S. Angelini J. G. Mcintosh M. Becker A. Rupp O. et al (2019). Genome Sequence of the Endophytic Strain Enterobacter Sp. J49, a Potential Biofertilizer for Peanut and Maize. Genomics111, 913–920. 10.1016/j.ygeno.2018.05.021
40
Mach K. J. Kraan C. M. Adger W. N. Buhaug H. Burke M. Fearon J. D. et al (2019). Climate as a Risk Factor for Armed Conflict. Nature571, 193–197. 10.1038/s41586-019-1300-6
41
Madhaiyan M. Poonguzhali S. Lee J.-S. Saravanan V. S. Lee K.-C. Santhanakrishnan P. (2010). Enterobacter Arachidis Sp. nov., a Plant-Growth-Promoting Diazotrophic Bacterium Isolated from Rhizosphere Soil of Groundnut. Int. J. Syst. Evol. Microbiol.60, 1559–1564. 10.1099/ijs.0.013664-0
42
Maheshwari R. Bhutani N. Suneja P. (2019). Screening and Characterization of Siderophore Producing Endophytic Bacteria from Cicer Arietinum and Pisum Sativum Plants. J. Appl. Biol. Biotechnol.7, 7–14. 10.7324/JABB.2019.70502
43
Majeed A. Abbasi M. K. Hameed S. Imran A. Rahim N. (2015). Isolation and Characterization of Plant Growth-Promoting Rhizobacteria from Wheat Rhizosphere and Their Effect on Plant Growth Promotion. Front. Microbiol.6, 198. 10.3389/fmicb.2015.00198
44
Masciarelli O. Llanes A. Luna V. (2014). A New PGPR Co-inoculated with Bradyrhizobium Japonicum Enhances Soybean Nodulation. Microbiol. Res.169, 609–615. 10.1016/j.micres.2013.10.001
45
Mclaggan D. Naprstek J. Buurman E. T. Epstein W. (1994). Interdependence of K+ and Glutamate Accumulation during Osmotic Adaptation of Escherichia coli. J. Biol. Chem.269, 1911–1917. 10.1016/s0021-9258(17)42113-2
46
Moriwaki T. Yamamoto Y. Aida T. Funahashi T. Shishido T. Asada M. et al (2008). Overexpression of the Escherichia coli Catalase Gene, katE, Enhances Tolerance to Salinity Stress in the Transgenic Indica Rice Cultivar, BR5. Plant Biotechnol. Rep.2, 41–46. 10.1007/s11816-008-0046-7
47
Mukherjee P. Roy P. (2016). Genomic Potential of Stenotrophomonas Maltophilia in Bioremediation with an Assessment of its Multifaceted Role in Our Environment. Front. Microbiol.7, 967. 10.3389/fmicb.2016.00967
48
Nascimento F. X. Hernandez A. G. Glick B. R. Rossi M. J. (2020a). The Extreme Plant‐growth‐promoting Properties of Pantoea Phytobeneficialis MSR2 Revealed by Functional and Genomic Analysis. Environ. Microbiol.22, 1341–1355. 10.1111/1462-2920.14946
49
Nascimento F. X. Hernández A. G. Glick B. R. Rossi M. J. (2020b). Plant Growth-Promoting Activities and Genomic Analysis of the Stress-Resistant Bacillus Megaterium STB1, a Bacterium of Agricultural and Biotechnological Interest. Biotechnol. Rep.25, e00406. 10.1016/j.btre.2019.e00406
50
Naveed M. Hussain M. B. Zahir Z. A. Mitter B. Sessitsch A. (2014). Drought Stress Amelioration in Wheat through Inoculation with Burkholderia Phytofirmans Strain PsJN. Plant Growth Regul.73, 121–131. 10.1007/s10725-013-9874-8
51
Ngumbi E. Kloepper J. (2016). Bacterial-mediated Drought Tolerance: Current and Future Prospects. Appl. Soil Ecol.105, 109–125. 10.1016/j.apsoil.2016.04.009
52
Nie L. Shah S. Rashid A. Burd G. I. Dixon D. G. Glick B. R. (2002). Phytoremediation of Arsenate Contaminated Soil by Transgenic Canola and the Plant Growth-Promoting Bacterium Enterobacter cloacae CAL2. Plant Physiology Biochem.40, 355–361. 10.1016/s0981-9428(02)01375-x
53
Nishijima K. Wall M. Siderhurst M. (2007). Demonstrating Pathogenicity of Enterobacter cloacae on Macadamia and Identifying Associated Volatiles of Gray Kernel of Macadamia in Hawaii. Plant Dis.91, 1221–1228. 10.1094/pdis-91-10-1221
54
Nurk S. Bankevich A. Antipov D. Gurevich A. A. Korobeynikov A. Lapidus A. et al (2013). Assembling Single-Cell Genomes and Mini-Metagenomes from Chimeric MDA Products. J. Comput. Biol.20, 714–737. 10.1089/cmb.2013.0084
55
Ojuederie O. B. Olanrewaju O. S. Babalola O. O. (2019). Plant Growth Promoting Rhizobacterial Mitigation of Drought Stress in Crop Plants: Implications for Sustainable Agriculture. Agronomy9, 712. 10.3390/agronomy9110712
56
Orozco-Mosqueda M. Flores A. Rojas-Sánchez B. Urtis-Flores C. A. Morales-Cedeño L. R. Valencia-Marin M. F. et al (2021). Plant Growth-Promoting Bacteria as Bioinoculants: Attributes and Challenges for Sustainable Crop Improvement. Agronomy11, 1167. 10.3390/agronomy11061167
57
Overbeek R. Olson R. Pusch G. D. Olsen G. J. Davis J. J. Disz T. et al (2014). The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res.42, D206–D214. 10.1093/nar/gkt1226
58
Pedraza R. O. Motok J. Salazar S. M. Ragout A. L. Mentel M. I. Tortora M. L. et al (2010). Growth-promotion of Strawberry Plants Inoculated with Azospirillum Brasilense. World J. Microbiol. Biotechnol.26, 265–272. 10.1007/s11274-009-0169-1
59
Peng G. Zhang W. Luo H. Xie H. Lai W. Tan Z. (2009). Enterobacter Oryzae Sp. nov., a Nitrogen-Fixing Bacterium Isolated from the Wild Rice Species Oryza Latifolia. Int. J. Syst. Evol. Microbiol.59, 1650–1655. 10.1099/ijs.0.005967-0
60
Ramachandran S. Fontanille P. Pandey A. Larroche C. (2006). Gluconic Acid: Properties, Applications and Microbial Production. Food Technol. Biotechnol.44, 185–195.
61
Ramakrishna W. Yadav R. Li K. (2019). Plant Growth Promoting Bacteria in Agriculture: Two Sides of a Coin. Appl. Soil Ecol.138, 10–18. 10.1016/j.apsoil.2019.02.019
62
Salehi-Lisar S. Y. Bakhshayeshan-Agdam H. (2016). “Drought Stress in Plants: Causes, Consequences, and Tolerance,” in Drought Stress Tolerance in Plants, Vol. 1. Editors HossainM.WaniS.BhattacharjeeS.BurrittD.TranL. S. (Cham: Springer), 1–16.
63
Samaras A. Nikolaidis M. Antequera-Gómez M. L. Cámara-Almirón J. Romero D. Moschakis T. et al (2021). Whole Genome Sequencing and Root Colonization Studies Reveal Novel Insights in the Biocontrol Potential and Growth Promotion by Bacillus Subtilis MBI 600 on Cucumber. Front. Microbiol.11, 600393. 10.3389/fmicb.2020.600393
64
Sandhya V. Sk Z A. Grover M. Reddy G. Venkateswarlu B. (2009). Alleviation of Drought Stress Effects in Sunflower Seedlings by the Exopolysaccharides Producing Pseudomonas Putida Strain GAP-P45. Biol. Fertil. Soils46, 17–26. 10.1007/s00374-009-0401-z
65
Santoyo G. Moreno-Hagelsieb G. Del Carmen Orozco-Mosqueda M. Glick B. R. (2016). Plant Growth-Promoting Bacterial Endophytes. Microbiol. Res.183, 92–99. 10.1016/j.micres.2015.11.008
66
Sati S. Bisht S. (2006). Utilization of Various Carbon Sources for the Growth of Waterborne Conidial Fungi. Mycologia98, 678–681. 10.3852/mycologia.98.5.678
67
Saum S. H. Sydow J. F. Palm P. Pfeiffer F. Oesterhelt D. Müller V. (2006). Biochemical and Molecular Characterization of the Biosynthesis of Glutamine and Glutamate, Two Major Compatible Solutes in the Moderately Halophilic Bacterium Halobacillus Halophilus. J. Bacteriol.188, 6808–6815. 10.1128/jb.00781-06
68
Schneider J. Wendisch V. F. (2011). Biotechnological Production of Polyamines by Bacteria: Recent Achievements and Future Perspectives. Appl. Microbiol. Biotechnol.91, 17–30. 10.1007/s00253-011-3252-0
69
Sha’arani S. Ramachandran V. Sabri N. S. A. Tahir A. A. Nur A. F. Akhir F. N. M. et al (2021). A Soil-Cooling Approach Supporting the Growth of Temperate Root Crops under a Tropical Climate. Trop. Agric. Dev.65, 146–152. 10.11248/jsta.65.146
70
Shah P. Swiatlo E. (2008). A Multifaceted Role for Polyamines in Bacterial Pathogens. Mol. Microbiol.68, 4–16. 10.1111/j.1365-2958.2008.06126.x
71
Shakeela S. Padder S. Bhat Z. (2017). Isolation and Characterization of Plant Growth Promoting Rhizobacteria Associated with Medicinal Plant Picrorhiza Kurroa. J. Pharmacogn. Phytochemistry6, 157–168.
72
Shariati J V. Malboobi M. A. Tabrizi Z. Tavakol E. Owlia P. Safari M. (2017). Comprehensive Genomic Analysis of a Plant Growth-Promoting Rhizobacterium Pantoea Agglomerans Strain P5. Sci. Rep.7, 15610–15612. 10.1038/s41598-017-15820-9
73
Spaepen S. Vanderleyden J. (2011). Auxin and Plant-Microbe Interactions. Cold Spring Harb. Perspect. Biol.3, a001438. 10.1101/cshperspect.a001438
74
Stein H. Honig A. Miller G. Erster O. Eilenberg H. Csonka L. N. et al (2011). Elevation of Free Proline and Proline-Rich Protein Levels by Simultaneous Manipulations of Proline Biosynthesis and Degradation in Plants. Plant Sci.181, 140–150. 10.1016/j.plantsci.2011.04.013
75
Strizhov N. Ábrahám E. Ökrész L. Blickling S. Zilberstein A. Schell J. et al (1997). Differential Expression of Two P5CS Genes Controlling Proline Accumulation during Salt‐stress Requires ABA and Is Regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J.12, 557–569. 10.1111/j.0960-7412.1997.00557.x
76
Tariq M. Hameed S. Yasmeen T. Zahid M. Zafar M. (2014). Molecular Characterization and Identification of Plant Growth Promoting Endophytic Bacteria Isolated from the Root Nodules of Pea (Pisum Sativum L). World J. Microbiol. Biotechnol.30, 719–725. 10.1007/s11274-013-1488-9
77
Timmusk S. Abd El-Daim I. A. Copolovici L. Tanilas T. Kännaste A. Behers L. et al (2014). Drought-tolerance of Wheat Improved by Rhizosphere Bacteria from Harsh Environments: Enhanced Biomass Production and Reduced Emissions of Stress Volatiles. PloS One9, e96086. 10.1371/journal.pone.0096086
78
Vinocur B. Altman A. (2005). Recent Advances in Engineering Plant Tolerance to Abiotic Stress: Achievements and Limitations. Curr. Opin. Biotechnol.16, 123–132. 10.1016/j.copbio.2005.02.001
79
Vurukonda S. S. K. P. Vardharajula S. Shrivastava M. Skz A. (2016). Enhancement of Drought Stress Tolerance in Crops by Plant Growth Promoting Rhizobacteria. Microbiol. Res.184, 13–24. 10.1016/j.micres.2015.12.003
80
Vurukonda S. S. K. P. Giovanardi D. Stefani E. (2018). Plant Growth Promoting and Biocontrol Activity of Streptomyces Spp. As Endophytes. Int. J. Mol. Sci.19, 952. 10.3390/ijms19040952
81
Wagh J. Shah S. Bhandari P. Archana G. Kumar G. N. (2014). Heterologous Expression of Pyrroloquinoline Quinone (Pqq) Gene Cluster Confers Mineral Phosphate Solubilization Ability to Herbaspirillum Seropedicae Z67. Appl. Microbiol. Biotechnol.98, 5117–5129. 10.1007/s00253-014-5610-1
82
Wang X. Preston J. F. 3rd Romeo T. (2004). The pgaABCD Locus of Escherichia coli Promotes the Synthesis of a Polysaccharide Adhesin Required for Biofilm Formation. J. Bacteriol.186, 2724–2734. 10.1128/jb.186.9.2724-2734.2004
83
Wani S. H. Kumar V. Shriram V. Sah S. K. (2016). Phytohormones and Their Metabolic Engineering for Abiotic Stress Tolerance in Crop Plants. Crop J.4, 162–176. 10.1016/j.cj.2016.01.010
84
Weber T. Blin K. Duddela S. Krug D. Kim H. U. Bruccoleri R. et al (2015). antiSMASH 3.0—a Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic acids Res.43, W237–W243. 10.1093/nar/gkv437
85
Zhu B. Lou M.-M. Xie G.-L. Wang G.-F. Zhou Q. Wang F. et al (2011). Enterobacter Mori Sp. nov., Associated with Bacterial Wilt on Morus Alba L. Int. J. Syst. Evol. Microbiol.61, 2769–2774. 10.1099/ijs.0.028613-0
Summary
Keywords
sustainable agriculture, abiotic stress, plant health improvement, rhizobacteria, crop productivity
Citation
Fadiji AE, Ayangbenro AS and Babalola OO (2023) Genomic Assessment of Enterobacter mori AYS9: A Potential Plant Growth-Promoting Drought-Resistant Rhizobacteria. Span. J. Soil Sci. 13:11302. doi: 10.3389/sjss.2023.11302
Received
23 February 2023
Accepted
31 May 2023
Published
08 June 2023
Volume
13 - 2023
Edited by
Avelino Núñez-Delgado, University of Santiago de Compostela, Spain
Updates
Copyright
© 2023 Fadiji, Ayangbenro and Babalola.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola Oluranti Babalola, olubukola.babalola@nwu.ac.za
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.