- 1Nephrology, Hemodialysis, Apheresis and Kidney Transplantation Department, University Hospital Grenoble, Grenoble, France
- 2Inserm U 1209, CNRS UMR 5309, Team Epigenetics, Immunity, Metabolism, Cell Signaling and Cancer, Institute for Advanced Biosciences Grenoble, University Grenoble Alpes, La Tronche, France
- 3Hepato-Gastroenterology and Digestive Oncology Department, University Hospital Grenoble, Grenoble, France
- 4Établissement Français du Sang Auvergne-Rhône-Alpes, R&D-Laboratory, Grenoble, France
- 5Translational Transplant Research Center, Icahn School of Medicine at Mount Sinai New York, New York, NY, United States
- 6Center for Transplantation Sciences, Department of Surgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
- 7Division of Nephrology, Department of Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
Immune cell metabolism plays a pivotal role in shaping and modulating immune responses. The metabolic state of immune cells influences their development, activation, differentiation, and overall function, impacting both innate and adaptive immunity. While glycolysis is crucial for activation and effector function of CD8 T cells, regulatory T cells mainly use oxidative phosphorylation and fatty acid oxidation, highlighting how different metabolic programs shape immune cells. Modification of cell metabolism may provide new therapeutic approaches to prevent rejection and avoid immunosuppressive toxicities. In particular, the distinct metabolic patterns of effector and suppressive cell subsets offer promising opportunities to target metabolic pathways that influence immune responses and graft outcomes. Herein, we review the main metabolic pathways used by immune cells, the techniques available to assay immune metabolism, and evidence supporting the possibility of shifting the immune response towards a tolerogenic profile by modifying energetic metabolism.
Introduction
For many decades, transplant immunology has focused on the mechanisms of organ rejection and developing strategies to prevent graft injury by blocking key activation pathways in the recipient’s immune system [1]. Changes in the metabolism of the alloimmune cells have been regarded as the downstream effect of their effector function. More recently, it has become apparent that changes in immune cell metabolism can, by themselves, drive immune cell fate. The advent of novel technologies has allowed the collection of detailed data to decipher the plasticity of the metabolic state of immune cells [2]. These findings highlight the metabolic pathways in immune cells as a potential novel therapeutic approach to reprogramming immune responses and preventing transplant rejection [3].
Herein, we review the current knowledge on the importance of metabolic changes in immune responses, recent technologies to study immune metabolism, and how targeting immune cell metabolism could improve outcomes in SOT recipients.
Cellular Metabolic Pathways in Immune Cells
Cellular metabolism is divided into anabolism and catabolism and both anabolic and catabolic reactions are essential for immune cell function and survival.
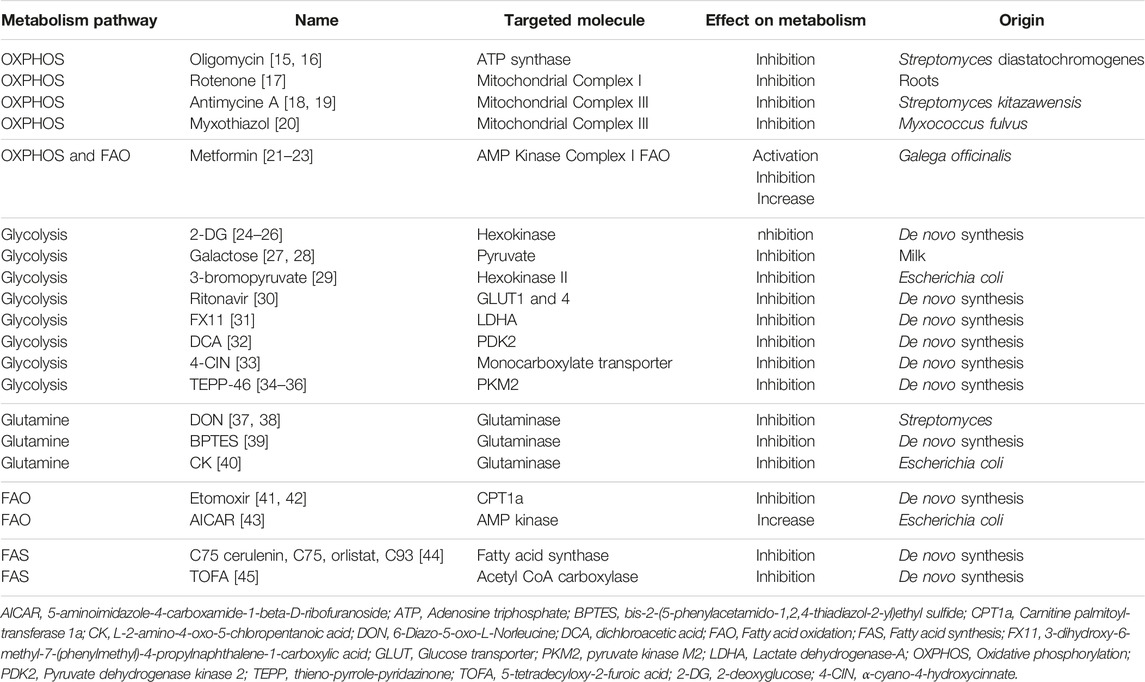
Anabolic reactions involve chain biosynthetic reactions that generates cell materials such as proteins and polypeptides from amino acids, DNA, RNA, lipids from fatty acid (FA). Anabolism require energy, typically provided in the form of adenosine triphosphate (ATP) molecules. Fatty acid synthesis (FAS) is a major anabolic reaction closely linked to immune cell function changes, differentiation, and proliferation [4]. Catabolic reactions involve the breakdown of complex molecules into simpler ones resulting in the release of energy such as proteins becoming amino acids or triglycerides breaking up into FA. Glycolysis and oxidative phosphorylation (OXPHOS) are the two main metabolic pathways that provide ATP for cells. Glycolysis refers to glucose oxidation to obtain ATP. OXPHOS refers to oxidation of nutriments within the mitochondria to generate ATP. Catabolic reactions are essential to support the high energetic requirements of immune cells, such as for cytokine production, rapid proliferation, and migratory activities (Figure 1).
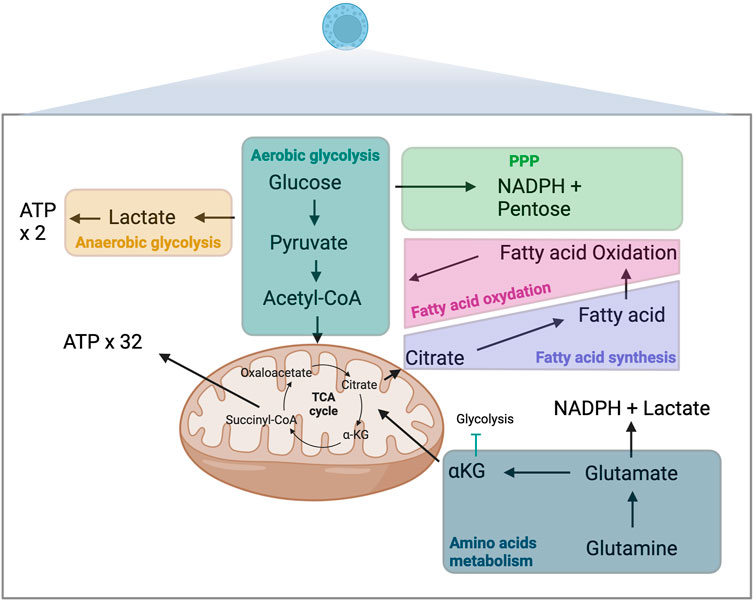
Figure 1. Metabolism and immune-metabolic pathways. ATP, adenosine triphosphate; αKG, α-ketoglutarate; TCA, tricarboxylic acid; NADPH, nicotinamide adenine dinucleotide phosphate; PPP, pentose phosphate pathway.
Sugars
Glycolysis, the breakdown of glucose, occurs in the cytosol of cells and is one of the primary catabolic processes contributing to the production of ATP [5]. The efficacy of the process depends not exclusively on oxygen availability and the mitochondrial capacity of immune cells. Aerobic glycolysis is the primary metabolic process contributing to energy generation in most immune cells. This highly efficient multi-step process starts with glucose molecule broken into two pyruvate molecules. In the presence of oxygen, which is required to re-oxidize nicotinamide adenine dinucleotide (NADH) to NAD+, pyruvate moves in the mitochondria and is converted to acetyl-CoA via pyruvate dehydrogenase. Acetyl-CoA enters in tricarboxylic acid (TCA) cycle and undergoes OXPHOS, leading to the production of 32 ATP molecules. In the absence of oxygen, glucose is metabolized in an anaerobic glycolysis process, through which pyruvate is converted into lactate, which yields only 2 ATP molecules. This process of lactate production can occur despite the presence of oxygen and fully functioning mitochondria (Warburg effect). Lactate also increases the NADH/NAD+ ratio. The pentose phosphate pathway (PPP) is an alternative pathway for glucose metabolism that generates nicotinamide adenine dinucleotide phosphate (NADPH) and pentoses (5-carbon sugars), essential moieties for synthesizing nucleotides. This pathway is crucial for effector functions of innate immune cells, including removing apoptotic cells including tolerogenic cells or activating and generating an oxidative burst of neutrophils [6, 7]. 5’ AMP-activated protein kinase (AMPK) is a metabolic sensor able to induce glycolysis trough the mammalian target of rapamycin complex (mTOR) pathway.
Amino acids
Glutamine is an essential substrate for immune cell metabolism. This non-essential amino acid can be transformed into glutamate and then into α-ketoglutarate (αKG), which like glucose-derived acetyl-CoA, is an essential fuel for the TCA cycle [8]. The second fate of glutamine-derived glutamate involves its transformation into lactate and NADPH trough a truncated TCA cycle in which succinyl-CoA is converted into succinate, fumarate and then malate [9]. Outside the mitochondria, malate will be converted into pyruvate and then lactate. Amino acids other than glutamine have also been shown to play essential roles in immune metabolism [10]. Tryptophane derived metabolites such as kynurenine or kynurenic acid have also emerged as a major pathway involved in regulatory T cells generation, in auto-immune diseases and in tolerance [11].
Fatty acids
Fatty acids (FAs) can fuel cellular metabolism through FA oxidation (FAO), another source of acetyl-CoA, which can then be shuttled to the TCA cycle. This metabolic pathway is of particular importance in adaptive immune responses [12]. AMPK is a metabolic sensor able to induce FAO.
Techniques to Study Immune Cell Metabolism
Different methods to measure cell metabolism have been used [13]. Each technique represents a different approach and has advantages and limitations [14]. Table 1 summarizes the available tools to block or activate the different metabolic pathways.
Global Oxygen and Acidification Measurement
For the last decade, OXPHOS and glycolysis have been measured using the Seahorse ® XF Analyzers (Figure 2A). This technique infers the oxygen consumption rate (OCR) through measuring the oxygen concentration in the supernatant of a cell culture over time, a surrogate marker of OXPHOS [46, 47]. Similarly, it estimates the extracellular acidification rate (ECAR) by measuring the changes in proton concentration in the supernatant over time, and using it as a surrogate marker of glycolysis [46]. OCR and ECAR can be assessed in parallel 96-well plate to perform replicates and multiple conditions.
Figure 2. Principal methods to assess metabolism in immune cells: measurement of oxygen (OCR) and the extracellular acidification rate (ECAR) in the supernatant (A), measurement of cell metabolism by single-cell energetic metabolism by profiling translation inhibition (SCENITH) (B), Cytometry Time Of Flight (C) and metabolomics assessment by mass spectrometry (D) OCR, oxygen consumption rate, ECAR, extracellular acidification rate.
This approach requires prior cell purification and it is unable to assess the metabolism at a single-cell level [48]. This method does not allow to perform simultaneous cell phenotyping nor cell sorting. Cells needs to be incubated from 12–18 h which can induce variability between wells in the number of cells and results.
Single-Cell Energetic Metabolism by Profiling Translation Inhibition (SCENITH)
The SCENITH method has recently provided an interesting additional tool to assess immune metabolism at a single-cell level using flow cytometry [49]. This method assumes that most of the cell’s energy is employed for protein synthesis [50, 51]. By using the ability of puromycin to incorporate into protein during synthesis and our ability to detect it with an anti-puromycin antibody, the SCENITH method utilizes the level of puromycin incorporation as a marker of protein synthesis and thus, a surrogate marker of cell global metabolic activity (Figure 2B). The advantages of this technique include the possibility to study metabolism at a single cell level, the simultaneous study of multiple cell subtypes, the lack of sensitivity to metabolic modifications induced by the media, and the requirement of only a low number of cells (∼2000 cells) without purification. An additional advantage is the possibility of assessing cell phenotyping and other functions concomitantly [52]. The main limitation of the SCENITH method is its reliance on protein synthesis, which is only an indirect marker of cell metabolism, and lacks relevance for cells with low levels of protein synthesis, such as quiescent cells.
Flow Cytometry and Cytometry by Time of Flight
By using a panel of key enzymes, flow cytometry can assess metabolic state. The “Met-Flow” panel includes 10 metabolic enzymes and transporters, including Hexokinase 1 for glycolysis, Carnitine palmitoyl-transferase 1A for FAO, Glucose-6-phosphate dehydrogenase [53]. This panel allows single-cell and phenotypic analysis of cell metabolism and does not require prior cell purification but the number of antibody needed may be a challenge.
Cytometry by time of flight (CyTOF) is another technique that can assess immune cell metabolism (Figure 2C). Instead of using fluorescent-labeled antibodies as in regular flow cytometry, cells are stained with antibodies conjugated to heavy metal isotopes [54], increasing the capacity to multiplex and reducing spectral overlap. About 110 metabolism-associated antibodies are available [55]. Recent studies reported a subset of 41–45 antibodies to target the important regulators (transporters, enzymes, signaling molecules, transcription factors) of metabolic pathways [56, 57]. CyTOF main limitation are the cost of the CyTOF equipment and the fact that this technique does not allow to recover living cells after analysis and thus only static measurement of single cells.
Metabolomics
Metabolomics encompasses methods to detect and measure the cell metabolite levels and modifications (mass spectrometry combined with chromatography or ion mass, protein weight, ionization, and magnetic resonance) [58]. Carbon-labeled tracers can be added to mass spectrometry to specifically interrogate metabolic enzyme activities. Isotope tracers allow to quantify metabolomic flux on top of metabolic concentrations [59]. Mass spectrometry allow to detect and quantify even low concentration metabolites. Metabolites are detected according to their mass and charge. Classical separation techniques are liquid or gas chromatography. These methods require the least amount of material (about 200 cells without purification) and can target specific metabolites or all the metabolome (Figure 2D). Because of unbiased analysis, it also allows the discovery of new or uncharacterized metabolites. Limitations of this technique are the potential impact of the handling (medium and storage) on metabolite levels, the impossibility of combining with phenotypic analyses, the cost of the equipment and the variability resulting of the metabolites quenching and purification that can change rapidly the level of metabolites.
How Metabolism Affects Immune Cells
Cellular metabolism does not only constitute a way to provide energy for immune cell survival and function, but it also regulates immune cell signaling pathways [60]. Metabolites have emerged as critical regulators of immune cell survival, differentiation, activation and function [5]. The metabolic network and its plasticity shape the fate and functions of both innate and adaptive immune cells [61].
Innate Immune Cells
Myeloid Cells: Dendritic Cells and Macrophages
Activation of DC through Toll-like receptors (TLRs) is a crucial step for DC activation, maturation, as well as antigen processing and presentation. TLR engagement is associated with an increased level of glycolysis and a decreased level of OXPHOS [62–64]. Interestingly, DC can switch to OXPHOS metabolism when deprived of glucose due to competitive glucose uptake by T cells in the context of antigen presentation and T cell activation. Notably, glucose deprivation increases the capacity of DC to present and stimulate T cells [65].
An increase in glycolysis is critical in the initial phase of DC proliferation and differentiation, but then specific inflammatory or tolerogenic metabolic reprogramming follows [66]. This has been illustrated through SCENITH and CyTOF analyses of DC metabolism which show an increase of AMPK pathway and a decrease of mTOR pathway in tolerogenic DC as compared to inflammatory DCs [67]. Tolerogenic DC have been show to highly increase the ECAR level in the presence of glucose, to produce more lactate and have a higher lactate dehydrogenase activity as compared to other DC, suggesting a strong glycolytic profile of those cells [68].
In addition, many studies demonstrate that tumors promote specific metabolic pathways and nutriment uptake as compared to innate immune cells to reduce their effector functions and escape immuno-surveillance [69].
Different metabolic profiles characterize macrophage subsets. Pro-inflammatory macrophages (M1) have a higher succinate dehydrogenase in the TCA cycle which results in an increase of succinate which stabilize Hypoxia Inducible Factor 1 α (HIF1α) and in turns, promotes and sustain glycolysis activity [70]. Conversely, anti-inflammatory macrophages (M2) exhibit enhanced FAO and OXPHOS activity with an intact TCA cycle. Interestingly, increasing glutamine concentration in vitro culture medium drives mouse macrophage polarization into M2 profile, proving support to the notion that it is possible to orient the immune response through metabolism modifications [71]. αKG, a product of glutaminolysis, acts as a sensor of pro-anti-inflammation signals in mouse macrophages and can promote M2 polarization, but the role of glutamine in human macrophage is unknown [71].
Natural Killer Cells
The NK cell metabolic profile and effector functions depend on the context and the microenvironment. Cytokine-driven NK cell activation is associated with increased mitochondrial OXPHOS and glycolysis [72]. The relationship between metabolism and NK cell function was shown in tumor models, in which reduced availability of glucose and amino acids (leucine, arginine, glutamine) results in NK cell function impairment [73, 74]. Tryptophane pathway induction by indole 2,3-diamine oxygen (IDO) in tumors results in NK cell apoptosis to promote survival of cancer cells [75].
T cells
Naive T cells
Metabolic program and T-cell activation are closely linked [8]. Before T Cell Receptor activation (TCR), naïve T cells are quiescent and have low ATP requirements. In their naïve state, their principal source of ATP is OXPHOS fueled by the oxidation of pyruvate and FAO, with a low glycolysis-based metabolism [8]. TCR engagement results in the activation, proliferation, and differentiation of the naïve T cells into effector, memory, and central memory T cells. This is paralleled by the transcription of key metabolic enzymes including the glucose transporter GLUT1 and the acetyl-CoA carboxylase 1 (ACC1) translation [76].
Activated T cells
T cell activation leads to major metabolic changes that favor glycolysis over OXPHOS [77].
Upon activation, CD8+ T cells undergo a first metabolic shift consisting of shuttling pyruvate to lactate metabolism [78], followed by a full switch from anaerobic to aerobic glycolysis [79]. The increase in glycolysis activity in activated CD8+ T cells is underpinned by an increase of glycolytic enzymes and an expression of glucose transporters such as GLUT1 [80]. Glycolysis inhibition results in cytokine and proliferation impairments in activated CD8+ T cells [81].
Effector CD4+ T cells, T helpers 1 (Th1), T helpers 2 (Th2), and T helpers 17 (Th17) cells are highly dependent on aerobic glycolysis, which is under HIF1α - mTOR regulation [82]. Although OXPHOS is more efficient in producing ATP, glycolysis gives the cells an advantage by rapidly providing the required energy for effector functions and proliferation. Interestingly, aerobic glycolysis is not needed for T cell activation, but it is strictly required for T effectors functions such as cytokine production (IL-2, IFN-γ mRNA translation and secretion) [77]. After TCR engagement and CD28 co-stimulation, the glucose uptake is increased by the upregulation of the cell surface glucose transporter GLUT-1 [83]. Interestingly, pharmacological blockade of glycolysis impairs Th1 and Th17 survival and function [84].
OXPHOS is also increased in activated CD4+ T cells even though the ratio OXPHOS/glycolysis is lower during T cell activation than in naïve CD4+T cells. The energy provided by OXPHOS seems to be mainly required during the first step of T cell activation, acting as an impulse [77]. CD4 T cells deficient for the mitochondrial complex III-derived Reactive oxygen Species (ROS) cannot activate and proliferate upon antigen presentation [85].
Amino acids are fundamental for activated T cells. Glutamine, a non-essential amino acid, constitutes an important energy source through glutaminolysis in activated T cells [86]. This is illustrated by the increase of glutamine transporters in activated T cells, and the reduced proliferation and cytokine secretion by T cells during glutamine starvation [87, 88]. Leucine have also been described to be of significant importance in the proliferation and differentiation of T cells trough [89].
T cell proliferation requires lipid synthesis to generate cell membranes for daughter cells. Blocking ACC1, a major enzyme for FAS, impairs Th1, Th2, and Th17 proliferation [90]. Interestingly, during Th17 cell development, but not regulatory T cells, FAS depends on ACC1, and blockade of this glycolytic-lipogenic pathway selectively impaired TH17 generation [90, 91].
Memory T cells
Memory T cells have a metabolic profile close to that of naïve T cells (lower glycolysis compared to OXPHOS) but with notable differences: they have a higher mitochondrial mass and a higher spare respiratory capacity which allow them to respond faster in case of antigen re-exposure [92]. Glycolysis in memory T cells is higher than in naïve T cells despite the similar ratio of glycolysis/OXPHOS [93]. As in regulatory T cells (Treg), FA constitute the principal fuel of OXPHOS for memory T cells [92]. FA come preferentially from de novo synthesis via the glycolytic-lipogenic pathway via mitochondrial citrate transformation and not from exogenous FA uptake, as in Tregs [94, 95].
AMPK is a regulator of FAO and glycolysis and the inability to generate memory T cells in AMPK-deficient mice is associated with deficient mitochondrial FAO [21]. Similarly, AMPK deficient CD8+ T cells are enable to generate memory CD8+ T cells [96–99]. In summary, metabolism signature of memory T cells remains uncertain while they exhibit an elevated profile of glycolysis and OXPHOS.
Exhausted T cells
In the context of persisting antigen and TCR stimulation, effector T cells progressively modify their phenotype to slowly become exhausted T cells, a low functional state phenotypically characterized by specific markers including programmed-death 1 (PD-1) [100]. In case of a high energy demand (glycolysis) sustained over time, glucose deprivation progressively drives a metabolic modification on effector T cells. These modifications are driven by the PD-1 pathway [101], resulting in a decrease in T cell glucose uptake and in OXPHOS [102], while FAO is upregulated [103].
Regulatory T cells
The metabolic profiles described above for other CD4+ T cells do not seem to apply to regulatory T cells (Treg), whose energy demands are not met through glycolysis but through OXPHOS and FAO [104].
Their independence from aerobic glycolysis has been shown in vivo in GLUT-1 deficient mice [105]. GLUT-1 deficiency was associated with impaired growth, proliferation, and survival of mature effector T cells, but did not affect either natural or induced Treg generation and expansion. The rate of glycolysis in Tregs is similar to that of naïve T cells but lower than in Th1 and Th17 cells [104, 105]. Inhibition of glycolysis using dichloroacetate increases Treg differentiation and promotes IL-10 production and FOXP3 expression [106]. Similarly, blocking glycolysis with 2-DG promotes Treg differentiation at the expense of Th17 [82].
In contrast, blocking OXPHOS results in Treg differentiation impairment [84]. In Treg, OXPHOS is fueled through FAO, blocking FAS has been shown to promote Treg generation, and FAO activity associates with an increase in AMPK activity. Adoptive transfer of modified OXPHOS or FAO deficient Tregs, resulted in a reduction of graft survival compared to wild-type [107]. Consistently, dysfunction of mitochondrial proteins (complex III, transcription factor A) is associated with Treg loss of function [108, 109].
B cells
Activation of B cells trough B cell receptor (BRC) increases glucose and amino-acid uptake [110, 111]. However, glucose is not used for glycolysis, but for PPP and nucleotide synthesis [112]. OXPHOS and TCA cycle are augmented in activated B cells, but they are fueled by other source of energy than glucose, such as FAs [112, 113].
Following antigen activation, naïve germinal center B cells migrate into the follicle, where somatic hypermutation and antibody affinity maturation occur. During this process, B cells display a significant increase in OXPHOS activation [114].
Thereafter, B cells are transformed into short-lived plasma cells outside the lymphoid follicle and then into long-lived plasma cells and memory B cells inside the follicle. In plasma cells, the production of antibodies requires a high production of glutamate from glutamine pathway and a lower rate of glycolysis [115]. However, T-dependent long-lived plasma cells are characterized by a higher glucose and amino-acid uptake as compared to short-lived plasma cells [116].
Immunometabolism in Solid Organ Transplantation
As the metabolism impacts the development and function of immune cells, alterations in metabolic pathways can modulate immune cell differentiation and subsequently affect the balance between pro-inflammatory and regulatory cells, and thus influence transplantation outcomes (Figure 3).
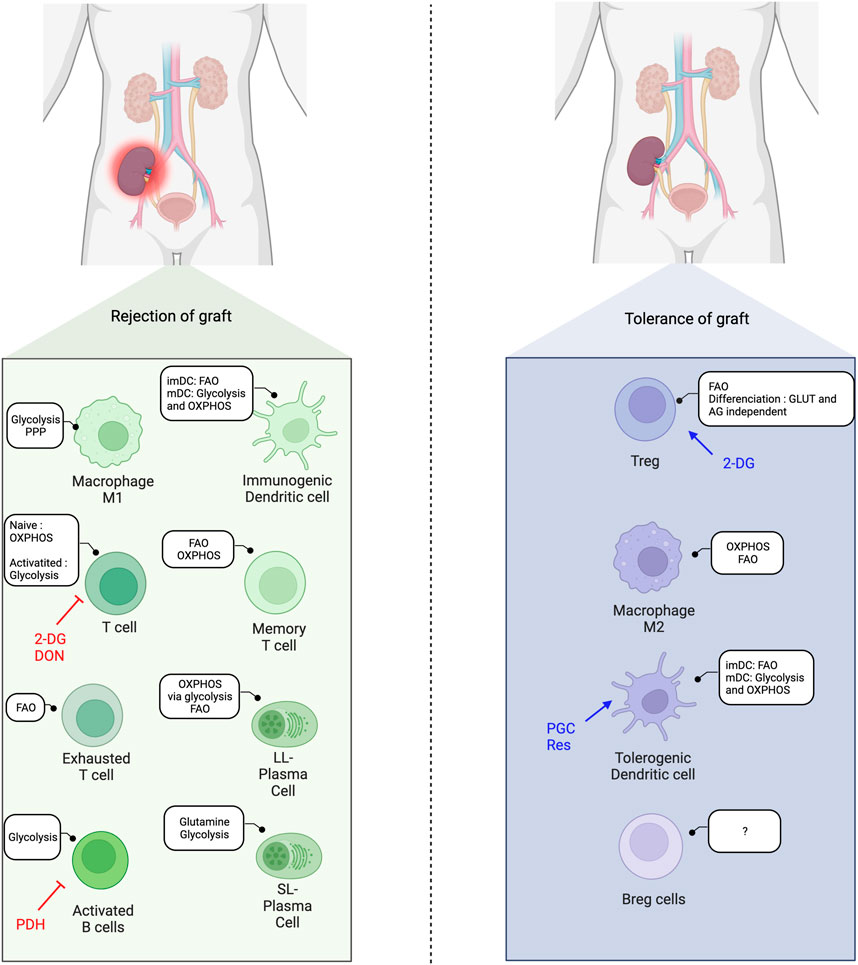
Figure 3. Immunometabolic balance in solid-organ transplantation and metabolic interventions. FAO, Fatty acid oxidation; OXPHOS, Oxidative phosphorylation; imDC, immature dendritic cells; mDC, mature dendritic cells; 2-DG, 2-deoxyglucose; DON, 6-Diazo-5-oxo-L-Norleucine; AG, aerobic glycolysis; Res, Resveatrol; PGC, PPARγ-coactivator-1β; PDH, pyruvate deshydrogenase; SL, short-lived; LL, long-lived.
Modulation of Immune Metabolism in SOT
Few but very promising studies in murine transplant models highlight the impact of metabolic reprogramming on the alloimmune response [14].
In 2015, Lee et al. were able to modulate the alloimmune response in a fully mismatched murine model of skin and heart allograft transplantation by targeting metabolic pathways [117]. The authors showed that glycolysis inhibition using 2-DG and metformin hindered proliferation and cytokine production in activated T cells. Combination of 2-DG with the glutamine inhibitor 6-diazo-5-oxo-l-norleucine (DON) resulted in an even more important inhibition of alloreactive CD4+ T cell proliferation and cytokine production, a decrease of acetyl-CoA levels and associated lipid synthesis, and a reduction of mTORC1 activation. The authors showed that in mice treated with 2-DG, metformin, and DON, CD4+ T cell kept their ability to differentiate into antigen-specific Foxp3+ CD4 T cells (Treg). Finally, the triple anti-metabolic therapy (2-DG, metformin, and DON), prolonged graft survival in a model of allogenic skin and heart transplantation, while discontinuation of treatment led to rapid graft rejection.
Immune metabolism may be modulated at the translational level. Quiescent CD4+ T cells accumulate a large amount of non-translated mRNA encoding key metabolic enzymes, which can be rapidly translated to activate aerobic glycolysis and FAO [76]. The engagement of the TCR triggers the translation of key proteins (GLUT1 and ACC1) for T cell activation and differentiation. Therefore, therapeutics that interfere with mRNA translation may affect alloimmunity by changing T cell metabolism.
Nian et al. showed that advanced age negatively impacts OXPHOS and glycolysis in naïve CD4+ T cells, and that glutaminolysis becomes the major source of ATP production later in life [118]. When the authors blocked OXPHOS with oligomycin, only CD4+ T cells from young mice were able to compensate for the metabolic loss with an increase in glycolysis. Interestingly, DON (glutaminolysis inhibitor) was able to inhibit IL-2, IFN-γ secretion, and cell proliferation in naïve CD4+ T cells from aged mice but not in naïve CD4+ T cells from young mice, which highlights the increasing reliance of naïve T cells on glutaminolysis with age. This approach prolonged graft survival and increased Tregs in a skin transplantation model in old mice. The results of DON on CD4+ T cells from aged mice were confirmed in human PBMC, suggesting its potential as a new age-dependent metabolic-mediated immunosuppression therapy [118].
Immune Metabolism Modulation in Combination With Costimulation Blockade
In 2020, Lee et al. used the combination of their triple anti-metabolic therapy (metformin, 2DG and DON) in association with a co-stimulatory blocker (CTLA4-Ig) [119]. Their model showed that CTLA4-Ig and metabolic inhibition have distinct but synergic effects on immune cells. They first showed that metabolic inhibition resulted in a higher inhibition of proliferation and promotion of apoptosis than CTLA4-Ig. Interestingly, in a model of skin allograft acute rejection in mice, they also showed prolonged graft survival with anti-metabolic drugs compared to CTLA4-Ig and controls. This may be explained by the costimulation-independent activation of memory T cells that CTLA4-Ig did not block. When metabolic inhibitors were added to CTLA4-Ig, there was an additive inhibiting effect on T cell proliferation, T-bet expression, and cytokine secretion. Finally, CTLA4-Ig and metabolic inhibition were synergic in preventing skin and heart allograft loss in their mice transplantation model.
Moreover, CTLA4-Ig addition to metabolic inhibitors allowed long-term acceptance of heart allograft, which was not possible when anti-metabolic therapy was given alone [117]. Priyadharshini et al. proposed that a sequential with first metabolism blockade (2DG) associated with CTLA4-Ig may induce tolerance phase that could be maintained by adding secondarily mTOR inhibitors. This strategy could specifically increase the Treg and tolerogenic DC in the context of SOT [120].
Conclusion
Recent studies and newly developed technologies have paved the way for modifying cell metabolism to influence the immune cell response. A better understanding of metabolic pathways in immune cells in the context of transplantation may offer the possibility to modulate the alloimmune response by reprogramming their metabolism to reshape specific immune cell subsets toward tolerogenic profiles. Further insights into metabolic dysregulation in SOT hold great promise to design novel therapies to improve graft and patient outcomes.
Author Contributions
JN wrote the first draft under ZM and PC’s guidance. CA, PM, LR, and MF provided comments and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
αKG, α-ketoglutarate; ACC1, acetyl-CoA carboxylase 1; AMPK, adenosine monophosphate–activated protein kinase; ATP, adenosine triphosphate; ECAR, extracellular acidification rate; FAO, fatty acid oxidation; HLA, human leukocyte antigen; IDO, indoleamine 2,3-dioxygenase; IFN, interferon; IL, interleukin; mTORC1, mammalian target of rapamycin complex 1; NADPH, nicotinamide adenine dinucleotide phosphate; OXPHOS, oxydative phosphorylation; OCR, oxygen consumption rate; PPP, pentose phosphate pathway; PBMC, peripheral blood mononuclear cells; SCENITH, single-cell energetic metabolism by profiling translation inhibition; SOT, solid-organ transplantation; TCA, tricarboxylic acid; TCR, T cell receptor; Tex, exhausted T cells; TLR, toll-like receptor; Treg, regulatory T cells.
References
1. Callemeyn, J, Lamarthée, B, Koenig, A, Koshy, P, Thaunat, O, and Naesens, M. Allorecognition and the Spectrum of Kidney Transplant Rejection. Kidney Int (2022) 101(4):692–710. doi:10.1016/j.kint.2021.11.029
2. Shapouri-Moghaddam, A, Mohammadian, S, Vazini, H, Taghadosi, M, Esmaeili, SA, Mardani, F, et al. Macrophage Plasticity, Polarization, and Function in Health and Disease. J Cel Physiol. (2018) 233(9):6425–40. doi:10.1002/jcp.26429
3. Zarrinpar, A, and Bensinger, SJ. The Therapeutic Potential of T Cell Metabolism. Am J Transpl (2017) 17(7):1705–12. doi:10.1111/ajt.14149
4. Qian, X, Yang, Z, Mao, E, and Chen, E. Regulation of Fatty Acid Synthesis in Immune Cells. Scand J Immunol (2018) 88(5):e12713. doi:10.1111/sji.12713
5. O’Neill, LAJ, Kishton, RJ, and Rathmell, J. A Guide to Immunometabolism for Immunologists. Nat Rev Immunol (2016) 16(9):553–65. doi:10.1038/nri.2016.70
6. Britt, EC, Lika, J, Giese, MA, Schoen, TJ, Seim, GL, Huang, Z, et al. Switching to the Cyclic Pentose Phosphate Pathway Powers the Oxidative Burst in Activated Neutrophils. Nat Metab (2022) 4(3):389–403. doi:10.1038/s42255-022-00550-8
7. He, D, Mao, Q, Jia, J, Wang, Z, Liu, Y, Liu, T, et al. Pentose Phosphate Pathway Regulates Tolerogenic Apoptotic Cell Clearance and Immune Tolerance. Front Immunol (2021) 12:797091. doi:10.3389/fimmu.2021.797091
8. Pearce, EL, Poffenberger, MC, Chang, CH, and Jones, RG. Fueling Immunity: Insights Into Metabolism and Lymphocyte Function. Science (2013) 342(6155):1242454. doi:10.1126/science.1242454
9. DeBerardinis, RJ, Mancuso, A, Daikhin, E, Nissim, I, Yudkoff, M, Wehrli, S, et al. Beyond Aerobic Glycolysis: Transformed Cells Can Engage in Glutamine Metabolism That Exceeds the Requirement for Protein and Nucleotide Synthesis. Proc Natl Acad Sci U S A. (2007) 104(49):19345–50. doi:10.1073/pnas.0709747104
10. Sinclair, LV, Rolf, J, Emslie, E, Shi, YB, Taylor, PM, and Cantrell, DA. Control of Amino-Acid Transport by Antigen Receptors Coordinates the Metabolic Reprogramming Essential for T Cell Differentiation. Nat Immunol. (2013) 14(5):500–8. doi:10.1038/ni.2556
11. Ding, X, Bin, P, Wu, W, Chang, Y, and Zhu, G. Tryptophan Metabolism, Regulatory T Cells, and Inflammatory Bowel Disease: A Mini Review. Mediators Inflamm (2020) 2020:9706140. doi:10.1155/2020/9706140
12. Zhou, X, Zhu, X, and Zeng, H. Fatty Acid Metabolism in Adaptive Immunity. FEBS J. (2023) 290(3):584–99. doi:10.1111/febs.16296
13. Voss, K, Hong, HS, Bader, JE, Sugiura, A, Lyssiotis, CA, and Rathmell, JC. A Guide to Interrogating Immunometabolism. Nat Rev Immunol (2021) 21(10):637–52. doi:10.1038/s41577-021-00529-8
14. Tanimine, N, Turka, LA, and Priyadharshini, B. Navigating T-Cell Immunometabolism in Transplantation. Transplantation (2018) 102(2):230–9. doi:10.1097/TP.0000000000001951
15. Devenish, RJ, Prescott, M, Boyle, GM, and Nagley, P. The Oligomycin Axis of Mitochondrial ATP Synthase: OSCP and the Proton Channel. J Bioenerg Biomembr. (2000) 32(5):507–15. doi:10.1023/a:1005621125812
16. Lardy, HA, Johnson, D, and McMURRAY, WC. Antibiotics as Tools for Metabolic Studies. I. A Survey of Toxic Antibiotics in Respiratory, Phosphorylative and Glycolytic Systems. Arch Biochem Biophys. (1958) 78(2):587–97. doi:10.1016/0003-9861(58)90383-7
17. Li, N, Ragheb, K, Lawler, G, Sturgis, J, Rajwa, B, Melendez, JA, et al. Mitochondrial Complex I Inhibitor Rotenone Induces Apoptosis Through Enhancing Mitochondrial Reactive Oxygen Species Production. J Biol Chem. (2003) 278(10):8516–25. doi:10.1074/jbc.M210432200
18. Nakayama, K, Okamoto, F, and HaradaAntimycin, YA. Antimycin A: Isolation From a New Streptomyces and Activity Against Rice Plant Blast Fungi. J Antibiot (Tokyo). (1956) 9(2):63–6.
19. Campo, ML, Kinnally, KW, and Tedeschi, H. The Effect of Antimycin A on Mouse Liver Inner Mitochondrial Membrane Channel Activity. J Biol Chem (1992) 267(12):8123–7. doi:10.1016/s0021-9258(18)42415-5
20. Thierbach, G, and Reichenbach, H. Myxothiazol, a New Inhibitor of the Cytochrome B-C1 Segment of Th Respiratory Chain. Biochim Biophys Acta. (1981) 638(2):282–9. doi:10.1016/0005-2728(81)90238-3
21. Pearce, EL, Walsh, MC, Cejas, PJ, Harms, GM, Shen, H, Wang, LS, et al. Enhancing CD8 T-Cell Memory by Modulating Fatty Acid Metabolism. Nature (2009) 460(7251):103–7. doi:10.1038/nature08097
22. Buzzai, M, Jones, RG, Amaravadi, RK, Lum, JJ, DeBerardinis, RJ, Zhao, F, et al. Systemic Treatment With the Antidiabetic Drug Metformin Selectively Impairs P53-Deficient Tumor Cell Growth. Cancer Res. (2007) 67(14):6745–52. doi:10.1158/0008-5472.CAN-06-4447
23. El-Mir, MY, Nogueira, V, Fontaine, E, Avéret, N, Rigoulet, M, and Leverve, X. Dimethylbiguanide Inhibits Cell Respiration via an Indirect Effect Targeted on the Respiratory Chain Complex I. J Biol Chem. (2000) 275(1):223–8. doi:10.1074/jbc.275.1.223
24. Rowe, I, Chiaravalli, M, Mannella, V, Ulisse, V, Quilici, G, Pema, M, et al. Defective Glucose Metabolism in Polycystic Kidney Disease Identifies a New Therapeutic Strategy. Nat Med. (2013) 19(4):488–93. doi:10.1038/nm.3092
25. Cheng, G, Zielonka, J, Dranka, BP, McAllister, D, Mackinnon, AC, Joseph, J, et al. Mitochondria-Targeted Drugs Synergize With 2-Deoxyglucose to Trigger Breast Cancer Cell Death. Cancer Res. (2012) 72(10):2634–44. doi:10.1158/0008-5472.CAN-11-3928
26. Dwarakanath, B, and Jain, V. Targeting Glucose Metabolism With 2-Deoxy-D-Glucose for Improving Cancer Therapy. Future Oncol. (2009) 5(5):581–5. doi:10.2217/fon.09.44
27. Robinson, BH, Petrova-Benedict, R, Buncic, JR, and Wallace, DC. Nonviability of Cells With Oxidative Defects in Galactose Medium: A Screening Test for Affected Patient Fibroblasts. Biochem Med Metab Biol. (1992) 48(2):122–6. doi:10.1016/0885-4505(92)90056-5
28. Marroquin, LD, Hynes, J, Dykens, JA, Jamieson, JD, and Will, Y. Circumventing the Crabtree Effect: Replacing Media Glucose With Galactose Increases Susceptibility of HepG2 Cells to Mitochondrial Toxicants. Toxicol Sci. (2007) 97(2):539–47. doi:10.1093/toxsci/kfm052
29. Lowe, PN, and Perham, RN. Bromopyruvate as an Active-Site-Directed Inhibitor of the Pyruvate Dehydrogenase Multienzyme Complex From Escherichia Coli. Biochemistry (1984) 23(1):91–7. doi:10.1021/bi00296a015
30. Hresko, RC, and Hruz, PW. HIV Protease Inhibitors Act as Competitive Inhibitors of the Cytoplasmic Glucose Binding Site of GLUTs With Differing Affinities for GLUT1 and GLUT4. PLoS One (2011) 6(9):e25237. doi:10.1371/journal.pone.0025237
31. Le, A, Cooper, CR, Gouw, AM, Dinavahi, R, Maitra, A, Deck, LM, et al. Inhibition of Lactate Dehydrogenase A Induces Oxidative Stress and Inhibits Tumor Progression. Proc Natl Acad Sci U S A. (2010) 107(5):2037–42. doi:10.1073/pnas.0914433107
32. Whitehouse, S, Cooper, RH, and Randle, PJ. Mechanism of Activation of Pyruvate Dehydrogenase by Dichloroacetate and Other Halogenated Carboxylic Acids. Biochem J. (1974) 141(3):761–74. doi:10.1042/bj1410761
33. Halestrap, AP, and Price, NT. The Proton-Linked Monocarboxylate Transporter (MCT) Family: Structure, Function and Regulation. Biochem J (1999) 343(Pt 2):281–99. doi:10.1042/bj3430281
34. Damasceno, LEA, Prado, DS, Veras, FP, Fonseca, MM, Toller-Kawahisa, JE, Rosa, MH, et al. PKM2 Promotes Th17 Cell Differentiation and Autoimmune Inflammation by Fine-Tuning STAT3 Activation. J Exp Med (2020) 217(10):e20190613. doi:10.1084/jem.20190613
35. Liu, C, Liu, C, and Fu, R. Research Progress on the Role of PKM2 in the Immune Response. Front Immunol (2022) 13:936967. doi:10.3389/fimmu.2022.936967
36. Angiari, S, Runtsch, MC, Sutton, CE, Palsson-McDermott, EM, Kelly, B, Rana, N, et al. Pharmacological Activation of Pyruvate Kinase M2 Inhibits CD4+ T Cell Pathogenicity and Suppresses Autoimmunity. Cell Metab. (2020) 31(2):391–405. doi:10.1016/j.cmet.2019.10.015
37. McElroy, KE, Bouchard, PJ, Harpel, MR, Horiuchi, KY, Rogers, KC, Murphy, DJ, et al. Implementation of a Continuous, Enzyme-Coupled Fluorescence Assay for High-Throughput Analysis of Glutamate-Producing Enzymes. Anal Biochem (2000) 284(2):382–7. doi:10.1006/abio.2000.4740
38. Thomas, AG, Rojas, C, Tanega, C, Shen, M, Simeonov, A, Boxer, MB, et al. Kinetic Characterization of Ebselen, Chelerythrine and Apomorphine as Glutaminase Inhibitors. Biochem Biophys Res Commun. (2013) 438(2):243–8. doi:10.1016/j.bbrc.2013.06.110
39. Robinson, MM, McBryant, SJ, Tsukamoto, T, Rojas, C, Ferraris, DV, Hamilton, SK, et al. Novel Mechanism of Inhibition of Rat Kidney-Type Glutaminase by Bis-2-(5-Phenylacetamido-1,2,4-Thiadiazol-2-Yl)ethyl Sulfide (BPTES). Biochem J (2007) 406(3):407–14. doi:10.1042/BJ20070039
40. Sekura, R, and Meister, A. Covalent Interaction of L-2-Amino-4-Oxo-5-Chloropentanoate at Glutamate Binding Site of Gamma-Glutamylcysteine Synthetase. J Biol Chem. (1977) 252(8):2606–10. doi:10.1016/s0021-9258(17)40501-1
41. Deberardinis, RJ, Lum, JJ, and Thompson, CB. Phosphatidylinositol 3-Kinase-Dependent Modulation of Carnitine Palmitoyltransferase 1A Expression Regulates Lipid Metabolism During Hematopoietic Cell Growth. J Biol Chem. (2006) 281(49):37372–80. doi:10.1074/jbc.M608372200
42. Lopaschuk, GD, Wall, SR, Olley, PM, and Davies, NJ. Etomoxir, a Carnitine Palmitoyltransferase I Inhibitor, Protects Hearts From Fatty Acid-Induced Ischemic Injury Independent of Changes in Long Chain Acylcarnitine. Circ Res. (1988) 63(6):1036–43. doi:10.1161/01.res.63.6.1036
43. Gaidhu, MP, Fediuc, S, and Ceddia, RB. 5-Aminoimidazole-4-Carboxamide-1-Beta-D-Ribofuranoside-Induced AMP-Activated Protein Kinase Phosphorylation Inhibits Basal and Insulin-Stimulated Glucose Uptake, Lipid Synthesis, and Fatty Acid Oxidation in Isolated Rat Adipocytes. J Biol Chem (2006) 281(36):25956–64. doi:10.1074/jbc.M602992200
44. Menendez, JA, and Lupu, R. Fatty Acid Synthase and the Lipogenic Phenotype in Cancer Pathogenesis. Nat Rev Cancer. (2007) 7(10):763–77. doi:10.1038/nrc2222
45. Wang, C, Xu, C, Sun, M, Luo, D, Liao, DF, and Cao, D. Acetyl-CoA Carboxylase-Alpha Inhibitor TOFA Induces Human Cancer Cell Apoptosis. Biochem Biophys Res Commun. (2009) 385(3):302–6. doi:10.1016/j.bbrc.2009.05.045
46. Plitzko, B, and Loesgen, S. Measurement of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in Culture Cells for Assessment of the Energy Metabolism. Bio Protoc. (2018) 8(10):e2850. doi:10.21769/BioProtoc.2850
47. Wu, M, Neilson, A, Swift, AL, Moran, R, Tamagnine, J, Parslow, D, et al. Multiparameter Metabolic Analysis Reveals a Close Link Between Attenuated Mitochondrial Bioenergetic Function and Enhanced Glycolysis Dependency in Human Tumor Cells. Am J Physiol Cell Physiol. (2007) 292(1):C125–36. doi:10.1152/ajpcell.00247.2006
48. Wei, Y, Ding, J, Li, J, Cai, S, Liu, S, Hong, L, et al. Metabolic Reprogramming of Immune Cells at the Maternal-Fetal Interface and the Development of Techniques for Immunometabolism. Front Immunol (2021) 12:717014. doi:10.3389/fimmu.2021.717014
49. Argüello, RJ, Combes, AJ, Char, R, Gigan, JP, Baaziz, AI, Bousiquot, E, et al. SCENITH: A Flow Cytometry-Based Method to Functionally Profile Energy Metabolism With Single-Cell Resolution. Cell Metab. (2020) 32(6):1063–75. doi:10.1016/j.cmet.2020.11.007
50. Buttgereit, F, and Brand, MD. A Hierarchy of ATP-Consuming Processes in Mammalian Cells. Biochem J (1995) 312:163–7. doi:10.1042/bj3120163
51. Lindqvist, LM, Tandoc, K, Topisirovic, I, and Furic, L. Cross-Talk Between Protein Synthesis, Energy Metabolism and Autophagy in Cancer. Curr Opin Genet Dev. (2018) 48:104–11. doi:10.1016/j.gde.2017.11.003
52. Artyomov, MN, and Van den Bossche, J. Immunometabolism in the Single-Cell Era. Cell Metab (2020) 32(5):710–25. doi:10.1016/j.cmet.2020.09.013
53. Ahl, PJ, Hopkins, RA, Xiang, WW, Au, B, Kaliaperumal, N, Fairhurst, AM, et al. Met-Flow, a Strategy for Single-Cell Metabolic Analysis Highlights Dynamic Changes in Immune Subpopulations. Commun Biol. (2020) 3(1):305. doi:10.1038/s42003-020-1027-9
54. Horowitz, A, Strauss-Albee, DM, Leipold, M, Kubo, J, Nemat-Gorgani, N, Dogan, OC, et al. Genetic and Environmental Determinants of Human NK Cell Diversity Revealed by Mass Cytometry. Sci Transl Med (2013) 23(208):208ra145. doi:10.1126/scitranslmed.3006702
55. Qian, L, and Guo, T. Immunometabolism the CyTOF Way. Immunity (2021) 54(4):610–3. doi:10.1016/j.immuni.2021.03.019
56. Hartmann, FJ, Mrdjen, D, McCaffrey, E, Glass, DR, Greenwald, NF, Bharadwaj, A, et al. Single-Cell Metabolic Profiling of Human Cytotoxic T Cells. Nat Biotechnol. (2021) 39(2):186–97. doi:10.1038/s41587-020-0651-8
57. Levine, LS. Single-Cell Metabolic Analysis by Mass Cytometry (2020). Available from: https://scholar.google.com/scholar_lookup?journal=BioRxiv&title=Single-cell+metabolic+analysis+by+mass+cytometry+reveals+distinct+transitional+states+of+CD8+T+cell+differentiation&author=LS+Levine&author=KJ+Hiam&author=DM+Marquez&author=I+Tenvooren&author=DC+Contreras&publication_year=2020& (Accessed January 14, 2024).
58. Tannahill, GM, Curtis, AM, Adamik, J, Palsson-McDermott, EM, McGettrick, AF, Goel, G, et al. Succinate Is an Inflammatory Signal That Induces IL-1β Through HIF-1α. Nature (2013) 496(7444):238–42. doi:10.1038/nature11986
59. Dong, W, Rawat, ES, Stephanopoulos, G, and Abu-Remaileh, M. Isotope Tracing in Health and Disease. Curr Opin Biotechnol. août (2022) 76:102739. doi:10.1016/j.copbio.2022.102739
60. Baker, SA, and Rutter, J. Metabolites as Signalling Molecules. Nat Rev Mol Cell Biol (2023) 24:355–74. doi:10.1038/s41580-022-00572-w
61. Hu, C, Xuan, Y, Zhang, X, Liu, Y, Yang, S, and Yang, K. Immune Cell Metabolism and Metabolic Reprogramming. Mol Biol Rep (2022) 49(10):9783–95. doi:10.1007/s11033-022-07474-2
62. Pantel, A, Teixeira, A, Haddad, E, Wood, EG, Steinman, RM, and Longhi, MP. Direct Type I IFN but Not MDA5/TLR3 Activation of Dendritic Cells Is Required for Maturation and Metabolic Shift to Glycolysis After Poly IC Stimulation. PLoS Biol. (2014) 12(1):e1001759. doi:10.1371/journal.pbio.1001759
63. Krawczyk, CM, Holowka, T, Sun, J, Blagih, J, Amiel, E, DeBerardinis, RJ, et al. Toll-Like Receptor-Induced Changes in Glycolytic Metabolism Regulate Dendritic Cell Activation. Blood (2010) 115(23):4742–9. doi:10.1182/blood-2009-10-249540
64. Pelgrom, LR, van der Ham, AJ, and Everts, B. Analysis of TLR-Induced Metabolic Changes in Dendritic Cells Using the Seahorse XF(e)96 Extracellular Flux Analyzer. Methods Mol Biol (2016) 1390:273–85. doi:10.1007/978-1-4939-3335-8_17
65. Lawless, SJ, Kedia-Mehta, N, Walls, JF, McGarrigle, R, Convery, O, Sinclair, LV, et al. Glucose Represses Dendritic Cell-Induced T Cell Responses. Nat Commun (2017) 8:15620. doi:10.1038/ncomms15620
66. Dong, H, and Bullock, TNJ. Metabolic Influences That Regulate Dendritic Cell Function in Tumors. Front Immunol (2014) 5:24. doi:10.3389/fimmu.2014.00024
67. Adamik, J, Munson, PV, Hartmann, FJ, Combes, AJ, Pierre, P, Krummel, MF, et al. Distinct Metabolic States Guide Maturation of Inflammatory and Tolerogenic Dendritic Cells. Nat Commun (2022) 13(1):5184. doi:10.1038/s41467-022-32849-1
68. Marin, E, Bouchet-Delbos, L, Renoult, O, Louvet, C, Nerriere-Daguin, V, Managh, AJ, et al. Human Tolerogenic Dendritic Cells Regulate Immune Responses Through Lactate Synthesis. Cell Metab. (2019) 30(6):1075–90. doi:10.1016/j.cmet.2019.11.011
69. Reinfeld, BI, Madden, MZ, Wolf, MM, Chytil, A, Bader, JE, Patterson, AR, et al. Cell-Programmed Nutrient Partitioning in the Tumour Microenvironment. Nature. mai (2021) 593(7858):282–8. doi:10.1038/s41586-021-03442-1
70. Viola, A, Munari, F, Sánchez-Rodríguez, R, Scolaro, T, and Castegna, A. The Metabolic Signature of Macrophage Responses. Front Immunol (2019) 10:1462. doi:10.3389/fimmu.2019.01462
71. Liu, PS, Wang, H, Li, X, Chao, T, Teav, T, Christen, S, et al. α-Ketoglutarate Orchestrates Macrophage Activation Through Metabolic and Epigenetic Reprogramming. Nat Immunol. (2017) 18(9):985–94. doi:10.1038/ni.3796
72. Terrén, I, Orrantia, A, Vitallé, J, Zenarruzabeitia, O, and Borrego, F. NK Cell Metabolism and Tumor Microenvironment. Front Immunol (2019) 10:2278. doi:10.3389/fimmu.2019.02278
73. Ohta, A. A Metabolic Immune Checkpoint: Adenosine in Tumor Microenvironment. Front Immunol (2016) 7:109. doi:10.3389/fimmu.2016.00109
74. Kedia-Mehta, N, and Finlay, DK. Competition for Nutrients and its Role in Controlling Immune Responses. Nat Commun. (2019) 10(1):2123. doi:10.1038/s41467-019-10015-4
75. Song, H, Park, H, Kim, YS, Kim, KD, Lee, HK, Cho, DH, et al. L-Kynurenine-Induced Apoptosis in Human NK Cells Is Mediated by Reactive Oxygen Species. Int Immunopharmacol. (2011) 11(8):932–8. doi:10.1016/j.intimp.2011.02.005
76. Ricciardi, S, Manfrini, N, Alfieri, R, Calamita, P, Crosti, MC, Gallo, S, et al. The Translational Machinery of Human CD4+ T Cells Is Poised for Activation and Controls the Switch From Quiescence to Metabolic Remodeling. Cell Metab. (2018) 28(6):895–906. doi:10.1016/j.cmet.2018.08.009
77. Chang, CH, Curtis, JD, Maggi, LB, Faubert, B, Villarino, AV, O’Sullivan, D, et al. Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell (2013) 153(6):1239–51. doi:10.1016/j.cell.2013.05.016
78. Pfeiffer, T, Schuster, S, and Bonhoeffer, S. Cooperation and Competition in the Evolution of ATP-Producing Pathways. Science (2001) 292(5516):504–7. doi:10.1126/science.1058079
79. Ma, EH, Verway, MJ, Johnson, RM, Roy, DG, Steadman, M, Hayes, S, et al. Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8+ T Cells. Immunity (2019) 51(5):856–70. doi:10.1016/j.immuni.2019.09.003
80. Menk, AV, Scharping, NE, Moreci, RS, Zeng, X, Guy, C, Salvatore, S, et al. Early TCR Signaling Induces Rapid Aerobic Glycolysis Enabling Distinct Acute T Cell Effector Functions. Cell Rep. (2018) 22(6):1509–21. doi:10.1016/j.celrep.2018.01.040
81. Cham, CM, Driessens, G, O’Keefe, JP, and Gajewski, TF. Glucose Deprivation Inhibits Multiple Key Gene Expression Events and Effector Functions in CD8+ T Cells. Eur J Immunol. (2008) 38(9):2438–50. doi:10.1002/eji.200838289
82. Shi, LZ, Wang, R, Huang, G, Vogel, P, Neale, G, Green, DR, et al. HIF1alpha-Dependent Glycolytic Pathway Orchestrates a Metabolic Checkpoint for the Differentiation of TH17 and Treg Cells. J Exp Med. (2011) 208(7):1367–76. doi:10.1084/jem.20110278
83. Jacobs, SR, Herman, CE, Maciver, NJ, Wofford, JA, Wieman, HL, Hammen, JJ, et al. Glucose Uptake Is Limiting in T Cell Activation and Requires CD28-Mediated Akt-Dependent and Independent Pathways. J Immunol. (2008) 180(7):4476–86. doi:10.4049/jimmunol.180.7.4476
84. Gerriets, VA, Kishton, RJ, Nichols, AG, Macintyre, AN, Inoue, M, Ilkayeva, O, et al. Metabolic Programming and PDHK1 Control CD4+ T Cell Subsets and Inflammation. J Clin Invest. (2015) 125(1):194–207. doi:10.1172/JCI76012
85. Sena, LA, Li, S, Jairaman, A, Prakriya, M, Ezponda, T, Hildeman, DA, et al. Mitochondria Are Required for Antigen-Specific T Cell Activation Through Reactive Oxygen Species Signaling. Immunity (2013) 38(2):225–36. doi:10.1016/j.immuni.2012.10.020
86. Kouidhi, S, Elgaaied, AB, and Chouaib, S. Impact of Metabolism on T-Cell Differentiation and Function and Cross Talk With Tumor Microenvironment. Front Immunol (2017) 8:270. doi:10.3389/fimmu.2017.00270
87. Carr, EL, Kelman, A, Wu, GS, Gopaul, R, Senkevitch, E, Aghvanyan, A, et al. Glutamine Uptake and Metabolism Are Coordinately Regulated by ERK/MAPK During T Lymphocyte Activation. J Immunol. (2010) 185(2):1037–44. doi:10.4049/jimmunol.0903586
88. Hammami, I, Chen, J, Bronte, V, DeCrescenzo, G, and Jolicoeur, M. L-Glutamine Is a Key Parameter in the Immunosuppression Phenomenon. Biochem Biophys Res Commun (2012) 425(4):724–9. doi:10.1016/j.bbrc.2012.07.139
89. Ananieva, EA, Powell, JD, and Hutson, SM. Leucine Metabolism in T Cell Activation: MTOR Signaling and Beyond. Adv Nutr. (2016) 7(4):798S–805S. doi:10.3945/an.115.011221
90. Berod, L, Friedrich, C, Nandan, A, Freitag, J, Hagemann, S, Harmrolfs, K, et al. De Novo Fatty Acid Synthesis Controls the Fate Between Regulatory T and T Helper 17 Cells. Nat Med. (2014) 20(11):1327–33. doi:10.1038/nm.3704
91. Tran, DT, Sundararaj, K, Atkinson, C, and Nadig, SN. T-Cell Immunometabolism: Therapeutic Implications in Organ Transplantation. Transplantation (2021) 105(11):e191–e201. doi:10.1097/TP.0000000000003767
92. van der Windt, GJW, Everts, B, Chang, CH, Curtis, JD, Freitas, TC, Amiel, E, et al. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity (2012) 36(1):68–78. doi:10.1016/j.immuni.2011.12.007
93. Gubser, PM, Bantug, GR, Razik, L, Fischer, M, Dimeloe, S, Hoenger, G, et al. Rapid Effector Function of Memory CD8+ T Cells Requires an Immediate-Early Glycolytic Switch. Nat Immunol (2013) 14(10):1064–72. doi:10.1038/ni.2687
94. O’Sullivan, D, van der Windt, GJW, Huang, SCC, Curtis, JD, Chang, CH, Buck, MD, et al. Memory CD8(+) T Cells Use Cell-Intrinsic Lipolysis to Support the Metabolic Programming Necessary for Development. Immunity (2014) 41(1):75–88. doi:10.1016/j.immuni.2014.06.005
95. Dufort, FJ, Gumina, MR, Ta, NL, Tao, Y, Heyse, SA, Scott, DA, et al. Glucose-Dependent De Novo Lipogenesis in B Lymphocytes: A Requirement for Atp-Citrate Lyase in Lipopolysaccharide-Induced Differentiation. J Biol Chem. (2014) 289(10):7011–24. doi:10.1074/jbc.M114.551051
96. Araki, K, Turner, AP, Shaffer, VO, Gangappa, S, Keller, SA, Bachmann, MF, et al. mTOR Regulates Memory CD8 T-Cell Differentiation. Nature (2009) 460(7251):108–12. doi:10.1038/nature08155
97. Tamás, P, Hawley, SA, Clarke, RG, Mustard, KJ, Green, K, Hardie, DG, et al. Regulation of the Energy Sensor AMP-Activated Protein Kinase by Antigen Receptor and Ca2+ in T Lymphocytes. J Exp Med (2006) 203(7):1665–70. doi:10.1084/jem.20052469
98. Rolf, J, Zarrouk, M, Finlay, DK, Foretz, M, Viollet, B, and Cantrell, DA. AMPKα1: A Glucose Sensor That Controls CD8 T-Cell Memory. Eur J Immunol. (2013) 43(4):889–96. doi:10.1002/eji.201243008
99. Ara, A, Xu, A, Ahmed, KA, Leary, SC, Islam, MF, Wu, Z, et al. The Energy Sensor AMPKα1 Is Critical in Rapamycin-Inhibition of mTORC1-S6k-Induced T-Cell Memory. Int J Mol Sci. (2021) 23(1):37. doi:10.3390/ijms23010037
100. Kulpa, DA, Lawani, M, Cooper, A, Peretz, Y, Ahlers, J, and Sékaly, RP. PD-1 Coinhibitory Signals: The Link Between Pathogenesis and Protection. Semin Immunol (2013) 25(3):219–27. doi:10.1016/j.smim.2013.02.002
101. Puigserver, P, and Spiegelman, BM. Peroxisome Proliferator-Activated Receptor-Gamma Coactivator 1 Alpha (PGC-1 Alpha): Transcriptional Coactivator and Metabolic Regulator. Endocr Rev. (2003) 24(1):78–90. doi:10.1210/er.2002-0012
102. Bengsch, B, Johnson, AL, Kurachi, M, Odorizzi, PM, Pauken, KE, Attanasio, J, et al. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8(+) T Cell Exhaustion. Immunity (2016) 45(2):358–73. doi:10.1016/j.immuni.2016.07.008
103. Patsoukis, N, Bardhan, K, Chatterjee, P, Sari, D, Liu, B, Bell, LN, et al. PD-1 Alters T-Cell Metabolic Reprogramming by Inhibiting Glycolysis and Promoting Lipolysis and Fatty Acid Oxidation. Nat Commun. (2015) 6:6692. doi:10.1038/ncomms7692
104. Michalek, RD, Gerriets, VA, Jacobs, SR, Macintyre, AN, MacIver, NJ, Mason, EF, et al. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4+ T Cell Subsets. J Immunol. (2011) 186(6):3299–303. doi:10.4049/jimmunol.1003613
105. Macintyre, AN, Gerriets, VA, Nichols, AG, Michalek, RD, Rudolph, MC, Deoliveira, D, et al. The Glucose Transporter Glut1 Is Selectively Essential for CD4 T Cell Activation and Effector Function. Cell Metab. (2014) 20(1):61–72. doi:10.1016/j.cmet.2014.05.004
106. Eleftheriadis, T, Pissas, G, Karioti, A, Antoniadi, G, Antoniadis, N, Liakopoulos, V, et al. Dichloroacetate at Therapeutic Concentration Alters Glucose Metabolism and Induces Regulatory T-Cell Differentiation in Alloreactive Human Lymphocytes. J Basic Clin Physiol Pharmacol (2013) 24(4):271–6. doi:10.1515/jbcpp-2013-0001
107. Beier, UH, Angelin, A, Akimova, T, Wang, L, Liu, Y, Xiao, H, et al. Essential Role of Mitochondrial Energy Metabolism in Foxp3+ T-Regulatory Cell Function and Allograft Survival. FASEB J. (2015) 29(6):2315–26.
108. Weinberg, SE, Singer, BD, Steinert, EM, Martinez, CA, Mehta, MM, Martínez-Reyes, I, et al. Mitochondrial Complex III Is Essential for Suppressive Function of Regulatory T Cells. Nature (2019) 565(7740):495–9. doi:10.1038/s41586-018-0846-z
109. Fu, Z, Ye, J, Dean, JW, Bostick, JW, Weinberg, SE, Xiong, L, et al. Requirement of Mitochondrial Transcription Factor A in Tissue-Resident Regulatory T Cell Maintenance and Function. Cell Rep (2019) 28(1):159–71. doi:10.1016/j.celrep.2019.06.024
110. Caro-Maldonado, A, Wang, R, Nichols, AG, Kuraoka, M, Milasta, S, Sun, LD, et al. Metabolic Reprogramming Is Required for Antibody Production That Is Suppressed in Anergic but Exaggerated in Chronically BAFF-Exposed B Cells. J Immunol. (2014) 192(8):3626–36. doi:10.4049/jimmunol.1302062
111. Cantor, J, Browne, CD, Ruppert, R, Féral, CC, Fässler, R, Rickert, RC, et al. CD98hc Facilitates B Cell Proliferation and Adaptive Humoral Immunity. Nat Immunol. (2009) 10(4):412–9. doi:10.1038/ni.1712
112. Waters, LR, Ahsan, FM, Wolf, DM, Shirihai, O, and Teitell, MA. Initial B Cell Activation Induces Metabolic Reprogramming and Mitochondrial Remodeling. iScience (2018) 5:99–109. doi:10.1016/j.isci.2018.07.005
113. Weisel, FJ, Mullett, SJ, Elsner, RA, Menk, AV, Trivedi, N, Luo, W, et al. Germinal Center B Cells Selectively Oxidize Fatty Acids for Energy While Conducting Minimal Glycolysis. Nat Immunol. (2020) 21(3):331–42. doi:10.1038/s41590-020-0598-4
114. Chen, D, Wang, Y, Manakkat Vijay, GK, Fu, S, Nash, CW, Xu, D, et al. Coupled Analysis of Transcriptome and BCR Mutations Reveals Role of OXPHOS in Affinity Maturation. Nat Immunol. (2021) 22(7):904–13. doi:10.1038/s41590-021-00936-y
115. Garcia-Manteiga, JM, Mari, S, Godejohann, M, Spraul, M, Napoli, C, Cenci, S, et al. Metabolomics of B to Plasma Cell Differentiation. J Proteome Res (2011) 10(9):4165–76. doi:10.1021/pr200328f
116. Lam, WY, Jash, A, Yao, CH, D’Souza, L, Wong, R, Nunley, RM, et al. Metabolic and Transcriptional Modules Independently Diversify Plasma Cell Lifespan and Function. Cell Rep (2018) 24(9):2479–92. doi:10.1016/j.celrep.2018.07.084
117. Lee, CF, Lo, YC, Cheng, CH, Furtmüller, GJ, Oh, B, Andrade-Oliveira, V, et al. Preventing Allograft Rejection by Targeting Immune Metabolism. Cell Rep (2015) 13(4):760–70. doi:10.1016/j.celrep.2015.09.036
118. Nian, Y, Iske, J, Maenosono, R, Minami, K, Heinbokel, T, Quante, M, et al. Targeting Age-Specific Changes in CD4+ T Cell Metabolism Ameliorates Alloimmune Responses and Prolongs Graft Survival. Aging Cell. (2021) 20(2):e13299. doi:10.1111/acel.13299
119. Cheng, CH, Lee, CF, Oh, BC, Furtmüller, GJ, Patel, CH, Brandacher, G, et al. Targeting Metabolism as a Platform for Inducing Allograft Tolerance in the Absence of Long-Term Immunosuppression. Front Immunol (2020) 11:572. doi:10.3389/fimmu.2020.00572
Keywords: solid organ transplantation, immune cells, metabolism, rejection, glycolysis
Citation: Noble J, Macek Jilkova Z, Aspord C, Malvezzi P, Fribourg M, Riella LV and Cravedi P (2024) Harnessing Immune Cell Metabolism to Modulate Alloresponse in Transplantation. Transpl Int 37:12330. doi: 10.3389/ti.2024.12330
Received: 29 October 2023; Accepted: 06 March 2024;
Published: 19 March 2024.
Copyright © 2024 Noble, Macek Jilkova, Aspord, Malvezzi, Fribourg, Riella and Cravedi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johan Noble, am5vYmxlQGNodS1ncmVub2JsZS5mcg==, am9oYW4ubm9ibGVAbXNzbS5lZHU=
†Present address:
Johan Noble, Translational Transplant Research Center, Icahn School of Medicine at Mount Sinai New York, New York, NY, United States
 Johan Noble
Johan Noble Zuzana Macek Jilkova2,3
Zuzana Macek Jilkova2,3 Paolo Malvezzi
Paolo Malvezzi Paolo Cravedi
Paolo Cravedi