In their recent article, Drs. Sablik and Sannier, along with more than 40 international collaborators, examined the impact of different microvascular inflammation (MVI) phenotypes on allograft outcomes by analyzing a total of 16,293 allograft biopsies from 6,798 patients across over 30 transplant centers in Europe and North America [1]. Clinical and pathological data was used to reclassify biopsy specimens according to the 2022 BANFF Classification of Renal Allograft Pathology now including the two new diagnostic categories of probable antibody-mediated rejection (ABMR) and MVI without evidence of an antibody-mediated response [2]. The newly identified phenotypes were present in 788 specimens, of which 641 were previously categorized as no rejection by the BANFF 2019 classification [3].
In terms of graft loss, patients with ABMR and those with the newly considered histopathological phenotype, MVI without antibody-mediated response (DSA-/C4d-) showed an increased risk of 2.7 (95% 2.2–3.3) and 2.1 (95% CI 1.5–3.1), respectively, when compared to non-rejection cases, whereas patients with the diagnosis of probable ABMR did not show an increased risk through the following 5 years after biopsy (Hazard Ratio [HR] of 1.3; 95% CI 0.8–2.1). In terms of progression to ABMR, patients with DSA-/C4d- MVI and those with probable ABMR showed a comparable risk of progression, with an intermediate cumulative incidence of ABMR during follow-up, positioned between patients without MVI and those with active ABMR (subdistribution HRs of 0.4 [95% CI, 0.3–0.5] and 0.7 [95% CI, 0.4–1.2], respectively). Finally, when analyzing the risk of progression to transplant glomerulopathy, the DSA-/C4d- MVI group showed, once more, a similar risk to that in the probable ABMR group, again falling in between the risks seen in those without MVI and those with active ABMR.
In short, this extensive population-based study, utilizing a remarkable dataset of allograft biopsies, compellingly demonstrates the importance of recognizing MVI as distinct histopathological phenotypes that associate with different disease progression and allograft failure. Notably, patients with MVI fulfilling the complete ABMR diagnosis display worse graft outcome, aligning with prior studies suggesting that patients with MVI and incomplete humoral phenotypes display better outcomes than those with full ABMR but worse than patients without rejection [4–6]. Crucially, the study emphasizes the necessity for broader acknowledgment of the MVI phenotypes in clinical practice, which have frequently been overlooked until recently. The discussed findings highlight the advantages of the 2022 BANFF classification in capturing the clinical, histological, and prognostic diversity of MVI over the previous version, thus establishing a foundation for standardizing future trials aimed at elucidating the immunological mechanisms behind these distinct phenotypes and potentially guiding tailored therapeutic strategies. Interestingly, the authors further propose that their findings may extend to other solid-organ transplants, where MVI is also a key diagnostic feature of ABMR, indicating possible similarities in pathophysiological processes that merit further study.
Authors are to be commended for their collaborative effort in assembling this substantial dataset to investigate the newly defined BANFF phenotypes in relation to the advent of distinct allograft outcomes. Nonetheless, the precise pathophysiological mechanisms underlying the development of these newly considered histopathological phenotypes, and especially MVI without evidence of an antibody-response (DSA-/C4d-) still remain elusive, leaving the question of what truly sets the spark for MVI. This is of paramount importance as the identification of main effector mechanisms orchestrating such specific graft injuries would allow to consistently design guided therapeutic strategies within interventional clinical trials. Indeed, a clear example underscoring such endeavor was delineated already in 2001, when the diagnostic feature of ABMR was first incorporated into the Banff classification by including the basic histopathological lesions of MVI and key immunological parameters such as serum DSA or C4d deposition [7], with an expansion of the histopathological ABMR criteria later in 2013 to include endarteritis, when concomitantly found in presence of serum DSA [8]. While the causality link between the two features may not strictly be confirmed, the strong associations described between such specific allograft lesions and the presence of DSA, the downstream effector mechanism of an anti-donor B-cell alloimmune response, has provided the solidest basis for this histological diagnosis. Notably, advances in molecular transcriptomics have helped to further refine distinct histopathological features, especially T-cell mediated rejection (TCMR) and ABMR, thus ultimately allowing reclassification of allograft lesions not fully captured with the conventional light microscope [9–13]. Nevertheless, while some recent works have shown overlapping transcriptional signatures between ABMR and DSA-/C4d- MVI, suggesting a common ethiopathological origin [14, 15], it may be argued that such common gene perturbation merely illustrates the similar cellular infiltrate composition, rather than the mechanisms driving its development. Notably, growing evidence suggests that DSA-/C4d- MVI may be more closely linked to an innate immune response, with natural killer (NK) cell–driven allorecognition potentially playing a key role in allograft injury [16–22]. Yet, the precise role of NK cells in MVI remains unclear [16], as they constitute only a small portion of the inflammatory infiltrate in MVI, which seems otherwise largely dominated by macrophages and T-cells [22–24]. Indeed, recent multi-omic profiling has shown a notable T-cell presence and activity, suggesting a T-cell effector dominant phenotype [25]. It is also plausible that other innate immune effector mechanisms, including myeloid-and monocyte-driven allorecognition could lead to similar histological/molecular pictures [26]. Importantly, it is highly likely that these diverse alloimmune effector mechanisms are not mutually exclusive but may, in fact, rather interconnect in complex ways [16].
Additionally, current clinical trials are exploring various blood- and urine-biomarkers indicative of graft injury, frequently caused by rejection, with donor-derived cell-free DNA (dd-cfDNA) emerging as particularly promising for differentiating microvascular injury in ABMR [27–38]. Consequently, dd-cfDNA has already been cleverly implemented into recent trials targeting ABMR. For instance, treatment with the anti-IL6 monoclonal antibody clazakizumab did not result in significant changes in dd-cfDNA levels, indicating ongoing allograft injury [39], whereas, only recently, treatment with anti-CD38 monoclonal antibody felzartamab demonstrated notable changes in dd-cfDNA, suggesting a beneficial therapeutic effect with the apparent resolution of injury [40]. Notably, while biomarkers like dd-cfDNA signal graft injury, they provide only limited insights into the underlying mechanisms driving graft damage [41]. Ideally, biomarkers would also reflect lesion pathophysiology or track alloimmune responses, allowing a more comprehensive understanding of the graft injury process. Thus, further research is needed to explore how biomarkers can aid in differentiating the various rejection phenotypes, understand rejection pathophysiology, assist in monitoring treatment responses, and be used to predict patient outcomes.
In consequence, as we now acknowledge novel kidney allograft rejection phenotypes and their different associated outcomes, it is essential to deepen our understanding of the main mechanisms driving these histopathological lesions by means of exploring immunological biomarkers and functional diagnostic tools tracking alloimmune responses, beyond conventional histology and DSA measurements. These advancements, along with others, will then represent a significant step forward in personalized care to optimize patient and allograft outcomes (Figure 1).
FIGURE 1
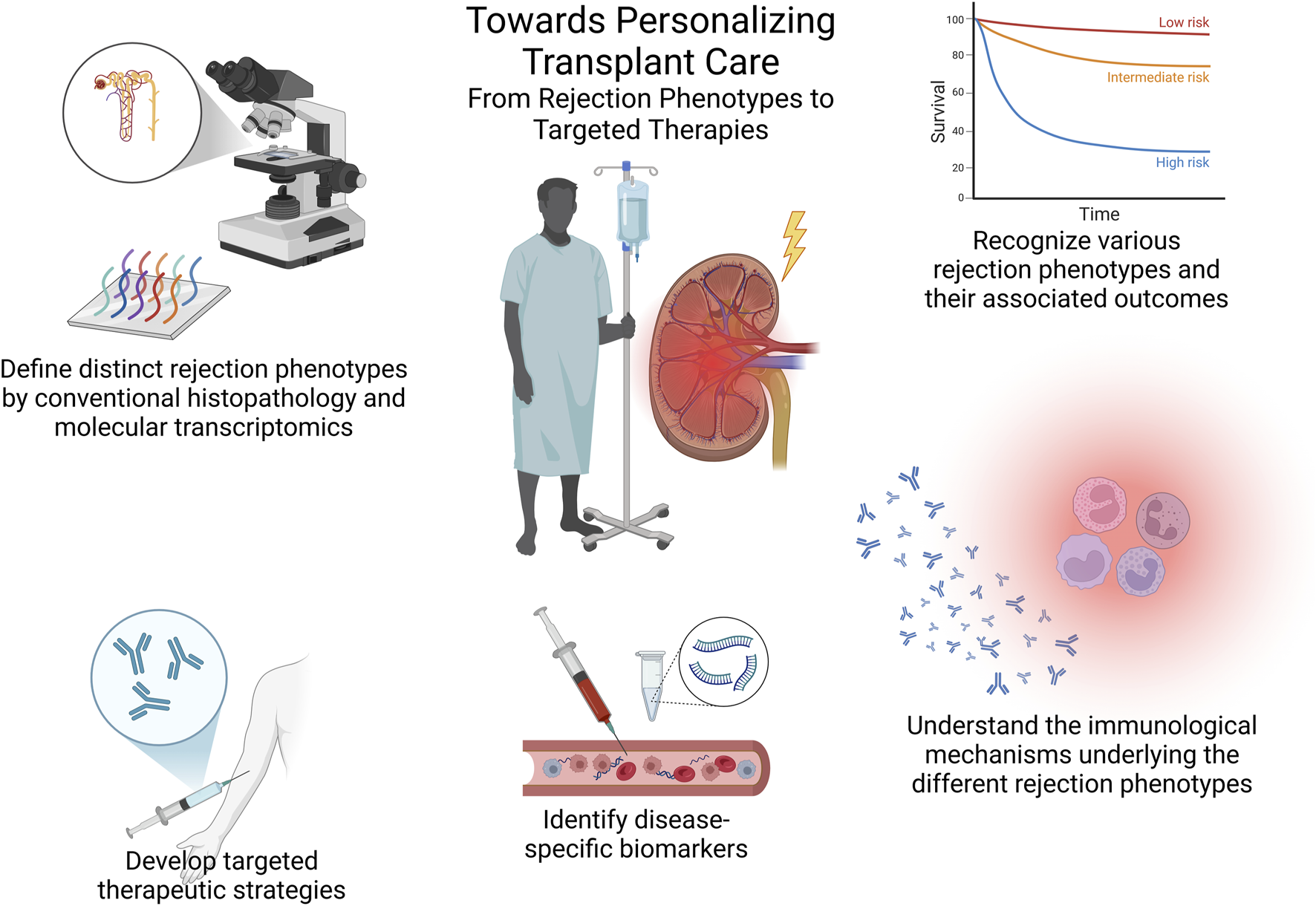
Towards personalizing transplant care – from rejection phenotypes to targeted therapies. This summary figure illustrates the essential steps toward personalized transplant care, including the classification of rejection phenotypes through conventional histopathology and molecular tools, understanding associated outcomes, exploring underlying immunological mechanisms, identifying disease-specific biomarkers, and ultimately developing targeted therapies based on these insights.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
LB and OB both drafted the article and revised it critically. Both authors contributed to the article and approved the submitted version.
Acknowledgments
The illustration in this article was made using Biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
References
1.
Sablik M Sannier A Raynaud M Goutaudier V Divard G Astor BC et al Microvascular Inflammation of Kidney Allografts and Clinical Outcomes. N Engl J Med (2024). 10.1056/nejmoa2408835
2.
Naesens M Roufosse C Haas M Lefaucheur C Mannon RB Adam BA et al The Banff 2022 Kidney Meeting Report: Reappraisal of Microvascular Inflammation and the Role of Biopsy-Based Transcript Diagnostics. Am J Transpl (2024) 24:338–49. 10.1016/j.ajt.2023.10.016
3.
Loupy A Haas M Roufosse C Naesens M Adam B Afrouzian M et al The Banff 2019 Kidney Meeting Report (I): Updates on and Clarification of Criteria for T Cell– and Antibody‐Mediated Rejection. Am J Transpl (2020) 20:2318–31. 10.1111/ajt.15898
4.
Senev A Coemans M Lerut E Sandt VV Daniëls L Kuypers D et al Histological Picture of Antibody-Mediated Rejection Without Donor-Specific Anti-HLA Antibodies: Clinical Presentation and Implications for Outcome. Am J Transpl (2019) 19:763–80. 10.1111/ajt.15074
5.
Callemeyn J Ameye H Lerut E Senev A Coemans M Loon EV et al Revisiting the Changes in the Banff Classification for Antibody-Mediated Rejection After Kidney Transplantation. Am J Transpl (2021) 21:2413–23. 10.1111/ajt.16474
6.
Buxeda A Llinàs-Mallol L Gimeno J Redondo-Pachón D Arias-Cabrales C Burballa C et al Microvascular Inflammation in the Absence of Human Leukocyte Antigen-Donor-Specific Antibody and C4d: An Orphan Category in Banff Classification With Cytotoxic T and Natural Killer Cell Infiltration. Am J Transpl (2023) 23:464–74. 10.1016/j.ajt.2022.12.018
7.
Racusen LC Colvin RB Solez K Mihatsch MJ Halloran PF Campbell PM et al Antibody-Mediated Rejection Criteria - An Addition to the Banff ’97 Classification of Renal Allograft Rejection. Am J Transpl (2003) 3:708–14. 10.1034/j.1600-6143.2003.00072.x
8.
Lefaucheur C Loupy A Vernerey D Duong-Van-Huyen J-P Suberbielle C Anglicheau D et al Antibody-Mediated Vascular Rejection of Kidney Allografts: A Population-Based Study. Lancet (2013) 381:313–9. 10.1016/s0140-6736(12)61265-3
9.
Halloran PF Reeve J Akalin E Aubert O Bohmig GA Brennan D et al Real Time Central Assessment of Kidney Transplant Indication Biopsies by Microarrays: The INTERCOMEX Study. Am J Transpl (2017) 17:2851–62. 10.1111/ajt.14329
10.
Halloran PF Matas A Kasiske BL Madill‐Thomsen KS Mackova M Famulski KS . Molecular Phenotype of Kidney Transplant Indication Biopsies With Inflammation in Scarred Areas. Am J Transpl (2019) 19:1356–70. 10.1111/ajt.15178
11.
Reeve J Böhmig GA Eskandary F Einecke G Lefaucheur C Loupy A et al Assessing Rejection-Related Disease in Kidney Transplant Biopsies Based on Archetypal Analysis of Molecular Phenotypes. JCI Insight (2017) 2:e94197. 10.1172/jci.insight.94197
12.
Madill‐Thomsen K Perkowska‐Ptasińska A Böhmig GA Eskandary F Einecke G Gupta G et al Discrepancy Analysis Comparing Molecular and Histology Diagnoses in Kidney Transplant Biopsies. Am J Transpl (2020) 20:1341–50. 10.1111/ajt.15752
13.
Garcia EC Giarraputo A Racapé M Goutaudier V Ursule-Dufait C Grange Pde la et al Antibody Mediated Rejection and T-Cell Mediated Rejection Molecular Signatures Using Next-Generation Sequencing in Kidney Transplant Biopsies. Transpl Int (2024) 37:13043. 10.3389/ti.2024.13043
14.
Halloran PF Madill-Thomsen KS Pon S Sikosana MLN Böhmig GA Bromberg J et al Molecular Diagnosis of ABMR With or Without Donor-Specific Antibody in Kidney Transplant Biopsies: Differences in Timing and Intensity But Similar Mechanisms and Outcomes. Am J Transpl (2022) 22:1976–91. 10.1111/ajt.17092
15.
Callemeyn J Lerut E Loor Hde Arijs I Thaunat O Koenig A et al Transcriptional Changes in Kidney Allografts With Histology of Antibody-Mediated Rejection Without Anti-HLA Donor-Specific Antibodies. J Am Soc Nephrol (2020) 31:2168–83. 10.1681/asn.2020030306
16.
Callemeyn J Lamarthée B Koenig A Koshy P Thaunat O Naesens M . Allorecognition and the Spectrum of Kidney Transplant Rejection. Kidney Int (2022) 101:692–710. 10.1016/j.kint.2021.11.029
17.
Koenig A Chen C-C Marçais A Barba T Mathias V Sicard A et al Missing Self Triggers NK Cell-Mediated Chronic Vascular Rejection of Solid Organ Transplants. Nat Commun (2019) 10:5350. 10.1038/s41467-019-13113-5
18.
Callemeyn J Senev A Coemans M Lerut E Sprangers B Kuypers D et al Missing Self–Induced Microvascular Rejection of Kidney Allografts: A Population-Based Study. J Am Soc Nephrol (2021) 32:2070–82. 10.1681/asn.2020111558
19.
Diebold M Vietzen H Heinzel A Haindl S Herz CT Mayer K et al Natural Killer Cell Functional Genetics and Donor-Specific Antibody-Triggered Microvascular Inflammation. Am J Transpl (2024) 24:743–54. 10.1016/j.ajt.2023.12.005
20.
Hidalgo LG Sis B Sellares J Campbell PM Mengel M Einecke G et al NK Cell Transcripts and NK Cells in Kidney Biopsies From Patients With Donor-Specific Antibodies: Evidence for NK Cell Involvement in Antibody-Mediated Rejection. Am J Transpl (2010) 10:1812–22. 10.1111/j.1600-6143.2010.03201.x
21.
Chambon M Koenig A . NK Cells: Not Just Followers But Also Initiators of Chronic Vascular Rejection. Transpl Int (2024) 37:13318. 10.3389/ti.2024.13318
22.
Yazdani S Callemeyn J Gazut S Lerut E Loor Hde Wevers M et al Natural Killer Cell Infiltration Is Discriminative for Antibody-Mediated Rejection and Predicts Outcome After Kidney Transplantation. Kidney Int (2019) 95:188–98. 10.1016/j.kint.2018.08.027
23.
Calvani J Terada M Lesaffre C Eloudzeri M Lamarthée B Burger C et al In situ Multiplex Immunofluorescence Analysis of the Inflammatory Burden in Kidney Allograft Rejection: A New Tool to Characterize the Alloimmune Response. Am J Transpl (2020) 20:942–53. 10.1111/ajt.15699
24.
Aguado-Domínguez E Cabrera-Pérez R Suarez-Benjumea A Abad-Molina C Núñez-Roldán A Aguilera I . Computer-Assisted Definition of the Inflammatory Infiltrates in Patients With Different Categories of Banff Kidney Allograft Rejection. Front Immunol (2019) 10:2605. 10.3389/fimmu.2019.02605
25.
Cristoferi I Varol H Baardwijk Mvan Rahiem L Lila KA Bosch TPPvan den et al Multiomic Profiling of Transplant Glomerulopathy Reveals a Novel T-Cell Dominant Subclass. Kidney Int (2024) 105:812–23. 10.1016/j.kint.2023.11.026
26.
Dai H Lan P Zhao D Abou-Daya K Liu W Chen W et al PIRs Mediate Innate Myeloid Cell Memory to Nonself MHC Molecules. Science (2020) 368:1122–7. 10.1126/science.aax4040
27.
Bloom RD Bromberg JS Poggio ED Bunnapradist S Langone AJ Sood P et al Cell-Free DNA and Active Rejection in Kidney Allografts. J Am Soc Nephrol (2017) 28:2221–32. 10.1681/asn.2016091034
28.
Halloran PF Reeve J Madill-Thomsen KS Demko Z Prewett A Billings P et al The Trifecta Study: Comparing Plasma Levels of Donor-Derived Cell-free DNA With the Molecular Phenotype of Kidney Transplant Biopsies. J Am Soc Nephrol (2022) 33:387–400. 10.1681/asn.2021091191
29.
Oellerich M Sherwood K Keown P Schütz E Beck J Stegbauer J et al Liquid Biopsies: Donor-Derived Cell-Free DNA for the Detection of Kidney Allograft Injury. Nat Rev Nephrol (2021) 17:591–603. 10.1038/s41581-021-00428-0
30.
Bu L Gupta G Pai A Anand S Stites E Moinuddin I et al Clinical Outcomes From the Assessing Donor-Derived Cell-Free DNA Monitoring Insights of Kidney Allografts With Longitudinal Surveillance (ADMIRAL) Study. Kidney Int (2022) 101:793–803. 10.1016/j.kint.2021.11.034
31.
Mayer KA Doberer K Tillgren A Viard T Haindl S Krivanec S et al Diagnostic Value of Donor‐Derived Cell‐Free DNA to Predict Antibody‐Mediated Rejection in Donor‐Specific Antibody‐Positive Renal Allograft Recipients. Transpl Int (2021) 34:1689–702. 10.1111/tri.13970
32.
Benning L Morath C Fink A Rudek M Speer C Kälble F et al Donor-Derived Cell-Free DNA (Dd-cfDNA) in Kidney Transplant Recipients With Indication Biopsy—Results of a Prospective Single-Center Trial. Transpl Int (2023) 36:11899. 10.3389/ti.2023.11899
33.
Jordan SC Bunnapradist S Bromberg JS Langone AJ Hiller D Yee JP et al Donor-Derived Cell-Free DNA Identifies Antibody-Mediated Rejection in Donor Specific Antibody Positive Kidney Transplant Recipients. Transpl Direct (2018) 4:e379. 10.1097/txd.0000000000000821
34.
Akifova A Budde K Oellerich M Beck J Bornemann-Kolatzki K Schütz E et al Perspective for Donor-Derived Cell-Free DNA in Antibody-Mediated Rejection After Kidney Transplantation: Defining Context of Use and Clinical Implications. Transpl Int (2024) 37:13239. 10.3389/ti.2024.13239
35.
Mantios E Filiopoulos V Constantoulakis P Liapis G Vittoraki A Casas S et al Assessment of Donor Derived Cell Free DNA (Dd-cfDNA) at Surveillance and at Clinical Suspicion of Acute Rejection in Renal Transplantation. Transpl Int (2023) 36:11507. 10.3389/ti.2023.11507
36.
Böhmer J Wasslavik C Andersson D Ståhlberg A Jonsson M Wåhlander H et al Absolute Quantification of Donor-Derived Cell-Free DNA in Pediatric and Adult Patients After Heart Transplantation: A Prospective Study. Transpl Int (2023) 36:11260. 10.3389/ti.2023.11260
37.
Kant S Brennan DC . Donor Derived Cell Free DNA in Kidney Transplantation: The Circa 2020–2021 Update. Transpl Int (2022) 35:10448. 10.3389/ti.2022.10448
38.
Verhoeven JGHP Hesselink DA Peeters AMA Jonge Ede Thüsen JHvon der Schaik RHNvan et al Donor-Derived Cell-Free DNA for the Detection of Heart Allograft Injury: The Impact of the Timing of the Liquid Biopsy. Transpl Int (2022) 35:10122. 10.3389/ti.2022.10122
39.
Mayer KA Doberer K Halloran PF Budde K Haindl S Mühlbacher J et al Anti-interleukin-6 Antibody Clazakizumab in Antibody-Mediated Kidney Transplant Rejection: Effect on Donor-Derived Cell-Free DNA and C-X-C Motif Chemokine Ligand 10. Transpl Direct (2022) 8:e1406. 10.1097/txd.0000000000001406
40.
Mayer KA Schrezenmeier E Diebold M Halloran PF Schatzl M Schranz S et al A Randomized Phase 2 Trial of Felzartamab in Antibody-Mediated Rejection. N Engl J Med (2024) 391:122–32. 10.1056/nejmoa2400763
41.
Pagliazzi A Bestard O Naesens M . Donor-Derived Cell-Free DNA: Attractive Biomarker Seeks a Context of Use. Transpl Int (2023) 36:12406. 10.3389/ti.2023.12406
Summary
Keywords
kidney transplantation, antibody-mediated rejection, biomarkers, microvascular inflammation, targeted therapies
Citation
Benning L and Bestard O (2024) Shedding Light on Microvascular Inflammation: Understanding Outcomes, But What Sparks the Flame?. Transpl Int 37:14032. doi: 10.3389/ti.2024.14032
Received
05 November 2024
Accepted
11 November 2024
Published
26 November 2024
Volume
37 - 2024
Updates
Copyright
© 2024 Benning and Bestard.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oriol Bestard, oriol.bestard@vallhebron.cat
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.