Abstract
Neuroblastoma is the most common extracranial solid tumor in children. Amplification of the MYCN gene has been observed in approximately 20%–30% of tumors. It is strongly correlated with advanced-stage disease, rapid tumor progression, resistance to chemotherapy and poor outcomes independent of patient age and stage of advanced disease. MYCN amplification identifies high-risk patients. To assess neuroblastoma tumors with MYCN amplification we used paraffin-embedded tissue sections in 57 patients and intraoperative tumor imprints in 10 patients by fluorescence in situ hybridization (FISH). Positive results for MYCN amplification have been observed in twelve patients’ paraffin-embedded tissue sections and in three patients’ intraoperative tumor imprints, which represents 22.4% of all patients tested in the analysis. Fluorescence in situ hybridization is a highly sensitive and useful technique for detecting MYCN amplification on paraffin-embedded tissue sections of neuroblastoma tumors and intraoperative tumor imprints thus facilitating therapeutic decisions based on the presence or absence of this important biologic marker. The presence of structural changes, regardless of MYCN gene amplification status, influences the clinical behavior of neuroblastoma. High-Density SNP Arrays have emerged as the perfect tools for detecting these changes due to their exceptional accuracy, sensitivity and ability to analyze copy number and allele information. Consequently, they are proven to be highly valuable in the genomic diagnosis of immature neuroectodermal tumors.
Introduction
Neuroblastoma is a malignant tumor in children aged 0–15 years. It accounts for 8%–10% of all childhood tumors, with 80% affecting children under the age of 5 years and approximately 15% representing all cancer deaths in children (Aygun, 2018; Swift et al., 2018). Approximately 60–70 of the new incidences of neuroblastoma are diagnosed each year in Poland (Whittle et al., 2017).
Neuroblastoma derives from primitive cells of the sympathetic nervous system. It occurs in the retroperitoneum, posterior mediastinum, neck, or pelvis. The clinical symptoms in neuroblastoma depend on the tumor mass, the extent of metastasis and catecholamine and vasoactive intestinal peptide secretion by the tumor cells. The diagnosis of neuroblastoma is based on histopathological results and the presence of tumor cells in the bone marrow accompanied by elevated levels of urinary catecholamines. In addition to clinical, histopathological and laboratory data, genetic test results are crucial in determining prognosis (Adamkiewicz-Drożyńska, 2006; Schulte et al., 2018).
The most common cytogenetic changes include deletions of chromosome 1p, amplification of the oncogene MYCN, ploidy changes, gains of chromosome arm 17q and deletion of 11q in tumor cells. Genetic abnormalities are very powerful predictors of response to therapy and outcome and provide important information to guide optimal patient management (Bhat and McGregor, 2017; Lim et al., 2020).
The MYCN gene is a cellular protooncogene of the MYC family. MYCN maps to the 2p24 chromosome. Structural alterations leading to amplification can manifest in the form of double minute chromosomes (dmins) or homogeneous staining regions (HSRs). Dmins represent extra chromosomal material, while HSRs denote duplicate material within chromosomes. Dmins are commonly observed in tumor-derived cells, whereas HSRs are more prevalent in cell lines. However, without traditional karyotype analysis, we are unable to precisely determine what type of changes we are dealing with (Van Noesel and Versteeg, 2004; Jeison et al., 2010).
MYCN is an oncogenic transcription factor involved in the regulation of proliferation, transformation, differentiation and apoptosis (Swift et al., 2018).
MYCN amplification is detected in 20%–30% of neuroblastoma patients (Yue et al., 2017; Otte et al., 2020). It is strongly correlated with advanced disease stage, rapid tumor progression, resistance to chemotherapy and poor outcomes. The MYCN amplification test result is included in medical protocols around the world, it is an independent indicator in the assessment of the patient outcome and it is one of the qualifying factors for a group requiring the most aggressive treatment (Brodeur et al., 1985; Huang and Weiss, 2013; Ahmed et al., 2017; Kaczówka et al., 2018; Croteau et al., 2021). MYCN amplification is present in 38% of patients with stages 3 and 4 (Ahmed et al., 2017) but only in 5%–10% of patients with stages 1, 2, and 4 s (Morandi, 2022). The level of MYCN gene amplification is necessary to avoid either under- or over-treating patients (Lee et al., 2018). The gold standard in the assessment of MYCN amplification is fluorescence in situ hybridization on paraffin-embedded tissue sections and intraoperative tumor imprints.
The independent factors that predict the outcome of treatment are: tumor histology, level of MYCN amplification and chromosomal copy number changes (Chicard et al., 2016; Lerone et al., 2021). In addition to these two main genetic markers, a number of segmental aberrations involving different chromosomes have also been described that, in addition to MYCN amplification, have a strong effect on the development of neuroblastoma, making it more aggressive and resistant to treatment (Lerone et al., 2021). Currently, the diagnosis of neuroblastoma also relies on molecular methods, such as microarrays, but it is not always possible to isolate good-quality DNA from paraffin-embedded tissue sections. An alternative method to obtain good-quality DNA is the intraoperative collection of tumor scrap from which DNA isolation is performed directly.
SNP array analysis provides comprehensive detection of the presence of deletions and duplications involving chromosomal fragments and allows for simultaneous assessment of DNA copy number.
SNP array analysis can also detect loss of heterozygosity (LOH). Studies of neuroblastoma cells have identified LOH in several chromosomal regions, of which LOH 1p and 4p correlate with poor patient outcomes.
In addition to these relatively large genomic aberrations, deletion aberrations of single genes, parts of genes or point mutations e.g., TERT, ALK, ATRX, and ARID1A, have recently been described as having unfavorable clinical outcomes and implications for clinical management (Brunner et al., 2016).
The aim of this study was to demonstrate the effectiveness of the FISH method for the evaluation of MYCN amplification on paraffin-embedded tissue sections and intraoperative tumor imprints together with HD SNP array analysis in neuroblastoma patients.
Materials and methods
The study materials consisted of 57 paraffin-embedded tissue sections with freshly removed tumors and 10 intraoperative tumor imprints, along with archival sections from 67 patients with an established clinical diagnosis of neuroblastoma (31 little girls and 36 little boys; median age: 3 years 2 months). All of the children were admitted to the Department of Pediatric Hematology, Oncology and Transplantology, Children’s University Hospital, Lublin, Poland between 1994 and 2023.
The collected clinical data included the patient`s age at diagnosis, stage according to the INSS (International Neuroblastoma Staging System) criteria (Park et al., 2017), tumor location, histopathological diagnosis of the examined tissues and MYCN amplification [Table 1]. Detailed patient data can be found in the Supplementary Material. All studies were performed between 1994-2023.
TABLE 1
| Criterium | Number | |
|---|---|---|
| Age at diagnosis | <18 m >18 m |
31 36 |
| Diagnosis | NB GNB GN ND |
52 11 1 3 |
| INSS risk group | 1 2 3 4s 4 |
7 3 24 3 30 |
| Primary site | Abdomen Adrenal glands Mediastinum Pelvis Cervical |
22 25 11 7 2 |
| MYCN amplification | Yes No |
15 52 |
Patient characteristics.
FISH method
We used the Pretreatment Kit (Vysis) and the LSI N-MYC (2p24) SpectrumGreen/CEP 2 SpectrumOrange Probe (Vysis) to estimate MYCN amplification.
Paraffin-embedded tissue blocks were cut into 4-micron thick sections on silanized slides and they were baked at 56°C overnight. Slides were soaked twice in xylene for 10 min and dehydrated in ethanol at 100%, 96%, 80%, and 70% for 2–5 min in each solution. Slides were then immersed in 0.2 N hydrochloric acid for 20 min and rinsed in water and wash buffer for 3 min in each of them. The slides were immersed in Pretreatment Solution at 80°C for 30 min and they were washed twice in 2xSSC for 5 min.
In the next step, the tissue sections were digested in protease solution at 37°C for 30 min and then the slides were immersed in 2xSSC cooled to 5°C.
The slides were then fixed in 10% formalin for 10 min. Fixed tissue, washed twice in 2xSSC and dried on a hot plate at 45°C–50°C.
Slides with intraoperative tumor imprints were immersed in a fixative prepared by mixing 3 parts methanol and 1 part acetic acid for 30 min. The slides were then dried at room temperature for 24 h.
To detect the presence of MYCN gene amplification, on paraffin-embedded tissue sections and intraoperative imprints of neuroblastoma tumors, we used fluorescence in situ hybridization (FISH) on interphase nuclei. Molecular cytogenetics uses the FISH method to identify specific DNA sequences on chromosomes using DNA probes that are directly labeled by the fluorophore. The labeled probe and the target DNA are denatured in the first step of this method. Hybridization is the second step of the FISH method. Combining the denatured probe and target DNA allows for the annealing of complementary DNA sequences.
We used 1 µL of direct fluorochrome-labeled, dual color DNA Probe for the MYCN gene locus (2p24) and α-satellite DNA identifying the centromeric region of chromosome 2 (Vysis LSI N-MYC (2p24) SpectrumGreen/CEP 2 SpectrumOrange Probe) combined with the 7 µL Hybridization Buffer and 2 µL water. The probe was placed on a slide, then covered with a coverslip and secured with rubber cement. The slides were incubated at 72°C for 5 min to denature the DNA. Hybridization was performed for 14–18 h at 37°C.
The slides were subsequently immersed in post-hybridization wash buffer at room temperature for 15–20 min until the coverslip fell off. Then they were immersed in post-hybridization wash buffer warmed to 72°C for 5 min. The cells were counterstained with DAPI. The signals were evaluated by eye or captured with Applied Spectral Imaging- ASI- Israel at ×100 magnification on the Nikon epifluorescence microscope equipped with a 100-W mercury arc lamp and an appropriate set of filters (single filters for FITC and TexasRed fluorochromes and a triple bandpass filter for FITC/TR/DAPI). A minimum of 200 nuclei was evaluated for each of the MYCN and CEP2 loci. Nuclei with two visible red and two green signals were scored as negative for MYCN amplification (Figure 1). According to the guidelines, the cutoff point for MYCN amplification was set at 10 signals (amplification ≥10; gain <10) only in the presence of disomy of chromosome 2. In the presence of polysomy of chromosome 2 in tumor cells, confirmation of MYCN gene amplification was based on the guideline that the number of tested signals must exceed that of control signals by at least four times (Ambros et al., 2009; Winnicka et al., 2020).
FIGURE 1
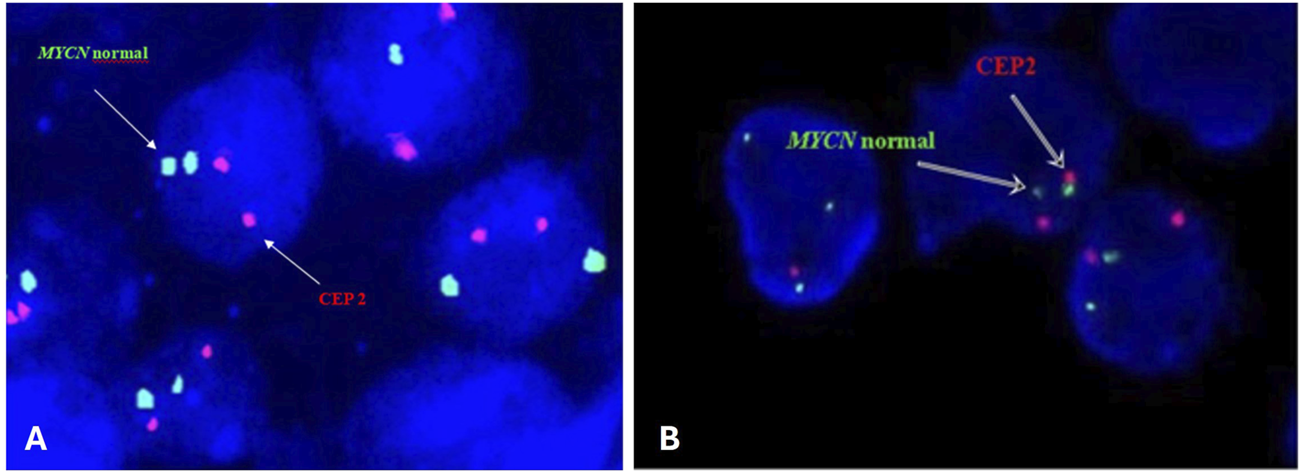
Representative FISH image on paraffin-embedded tissue section (A) and intraoperative tumor imprints (B) using the Vysis LSI N-MYC (2p24) SpectrumGreen/CEP 2 SpectrumOrange Probe from a neuroblastoma tumor showing normal interphase nuclei captured with Applied Spectral Imaging- ASI-Israel at ×100 magnification on the Nikon epifluorescence microscope.
The study was approved by the Ethics Committee of the Medical University of Lublin (KE-0254/229/2012).
HD SNP array method
In eight patients, we isolated DNA from tumor tissue and performed microarray testing using the CytoScan HD array to evaluate additional cytogenetic abnormalities. In one case, the quality of the DNA did not allow for the array procedure.
Genomic DNA was isolated with the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols. DNA samples were stored at −20°C until the next step. The concentration and quality of DNA were determined using a spectrophotometric method (NanoDrop 8000; Thermo Fisher Scientific, Waltham, MA, United States). In total, 250 ng of genomic DNA was used in accordance with the manufacturer’s protocols. Microarray testing was performed with the use of a CytoScan HD array [2,670,000 markers, including 750,000 SNP and 1,900,000 non-polymorphic copy number variant (CNV) markers] (Applied Biosystems, part of Thermo Fisher Scientific). All subsequent steps and data analysis were performed as described previously by Lejman et al. (2020). The scanned data file was analyzed with Chromosome Analysis Suite v 3.3 (ChAS; Thermo Fisher Scientific) using the CRCh37 (hg19) reference genome.
Results
Results of MYCN amplification by fluorescence in situ hybridization (FISH)
Sixty-seven patients participated in this study. Thirty-four (50.7%) of patients had localized neuroblastoma (1–3 stages), 4.5% stage 4S, and 44.8% stage 4 disease. Forty-seven (70.1%) of patients had an abdominal/adrenal mass at diagnosis. Thirty-one (46.3%) patients were <18 months of age, and thirty-six (53.7%) patients were >18 months of age. Of 67 patient samples, including 52 neuroblastomas, 11 ganglioneuroblastomas, and 1 ganglioneuroma, 3 samples had no definitive diagnosis.
Positive results for MYCN amplification were observed in twelve patients’ paraffin-embedded tissue sections and in three patients’ intraoperative tumor imprints which represents 22.4% of all tested patients in the analysis (Figure 2).
FIGURE 2
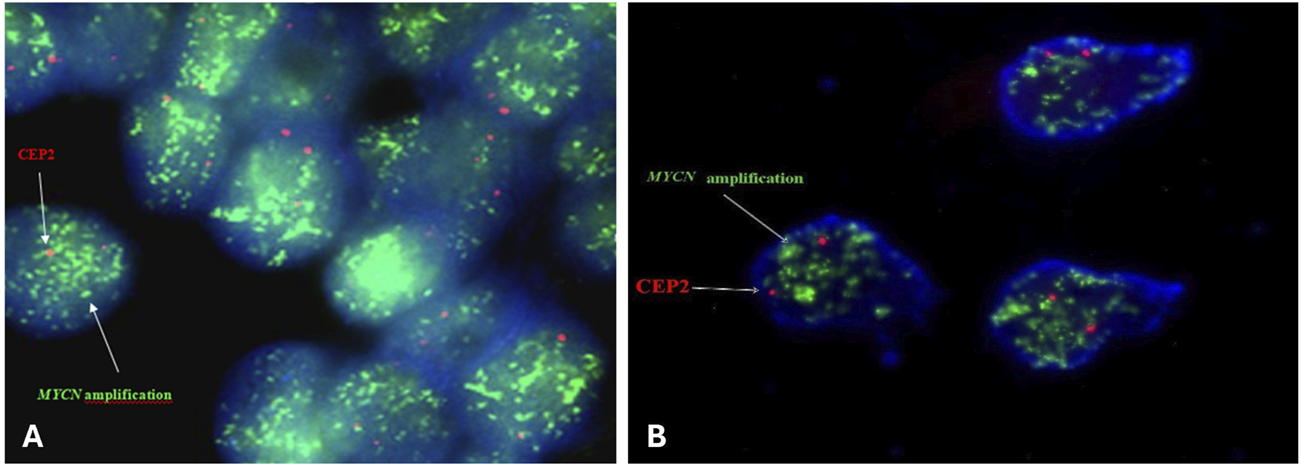
Representative FISH image on paraffin-embedded tissue section (A) and intraoperative tumor imprints (B) using Vysis LSI N-MYC (2p24) SpectrumGreen/CEP 2 SpectrumOrange Probe from a neuroblastoma tumor showing amplification MYCN captured with Applied Spectral Imaging- ASI- Israel at ×100 magnification on the Nikon epifluorescence microscope.
There were 9 (60%) little boys and 6 (40%) little girls included in the study. Of the 15 patients, 1 (6.7%) had stage 3 disease, 1 (6.7%) had stage 4 s disease, and 13 (86.6%) had stage 4 disease. Of these 15 patients with MYCN amplifications 5 (33.3%) were <18 months of age, and 10 (66.7%) were >18 months of age. Five (33.3%) of the patients with MYCN amplification had a tumor mass in the abdomen and ten (66.7%) had a tumor mass in the adrenal gland.
Molecular karyotype results
To evaluate additional molecular abnormalities we performed microarray testing on 6 patients with different stages of disease, tumor localization and ages at diagnosis.
We performed microarray testing on a patient with stage 4 disease and tumor localization in the adrenal gland. The patient was a girl who was over 18 months of age at the time of diagnosis. Through fluorescence in situ hybridization we observed positive results for MYCN amplification. Microarray analysis revealed a deletion within the short arm of chromosome 1 (arr [GRCh37] 1p36.33p36.22 (849466_9304205)x1), and a duplication involving a fragment of the short arm of chromosome 1, and the long arm of chromosome 1 (arr [GRCh37] 1p13.3q44 (110062169_249224684)x3). In region 2p25.3p21 we observed chromothripsis (arr [GRCh37] 2p25.3p21 (12770_46642249)cth) (Figure 3). The MYCN gene (2p24.3) was amplified in this region (10 copies). In addition, the ALK gene (2p23.2-p23.1) was also duplicated. Monosomy of chromosome 3 and trisomy of chromosome 18 were identified (Figure 4).
FIGURE 3
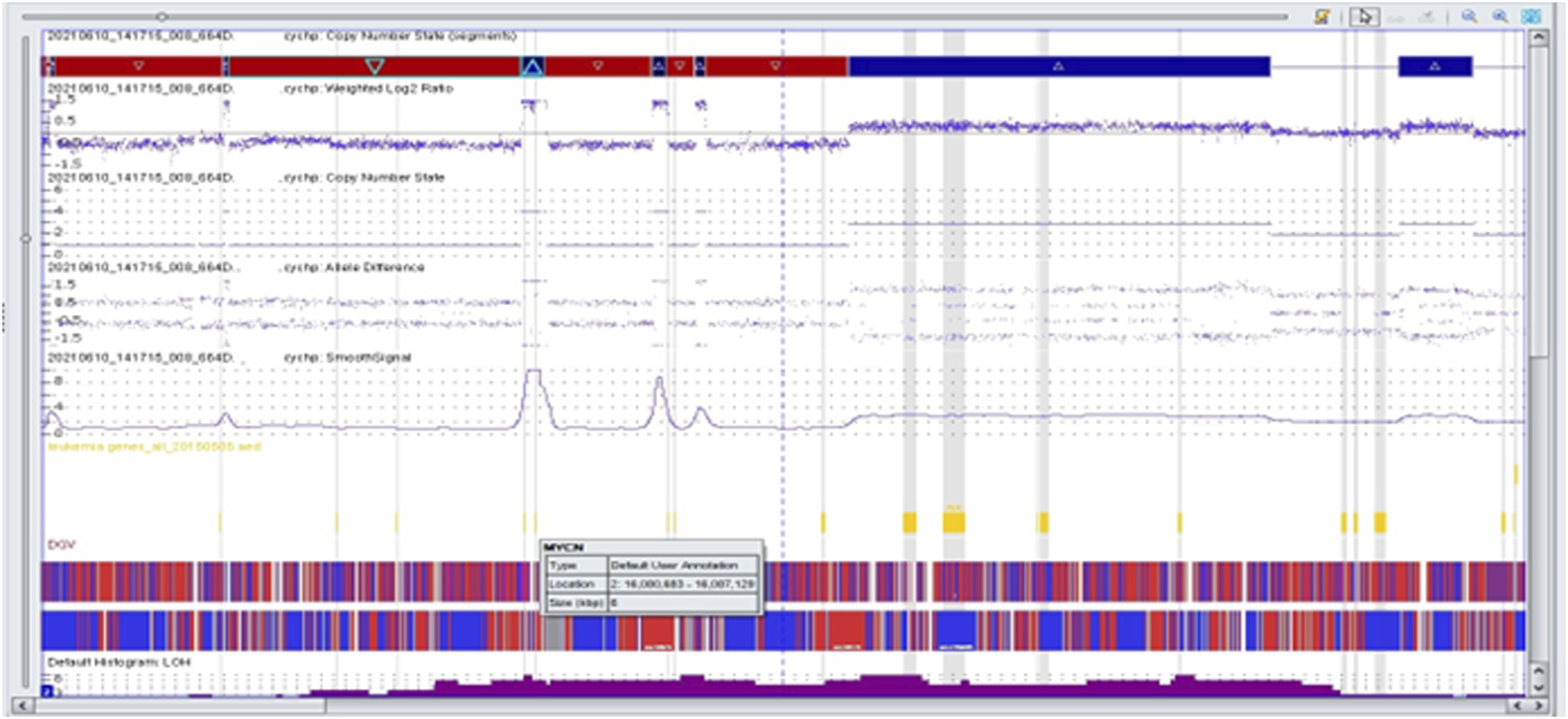
Complex copy number gains and losses in the region 2p25.3p21 of chromosome 2 in patient 63. With Neuroblastoma by single-nucleotide polymorphism (SNP) array. Colored segmental regions (top line: red color—deletions; blue color—gains), copy number status, allelic difference, and smooth signal aberrations by SNP array.
FIGURE 4
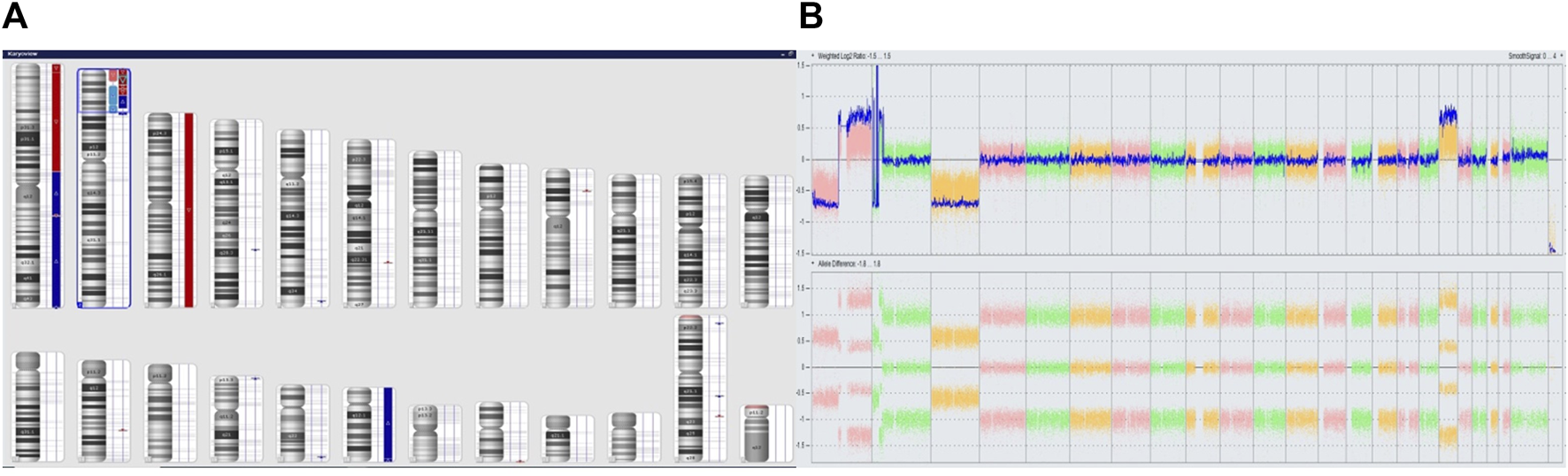
Complex copy number gains and losses in the whole genome of patient 63. With Neuroblastoma. Colored segmental regions (top line: red color—deletions; blue color—gains) (A) and Whole Genome View (B).
The second patient was a boy over 18 months of age at diagnosis, with stage 4 disease and tumor localization in the adrenal gland. Through fluorescence in situ hybridization we did not observe any amplification of the MYCN gene. Microarray analysis identified 3 cell lines with various genetic changes (Figure 5). We present the results in the table below (Table 2).
FIGURE 5
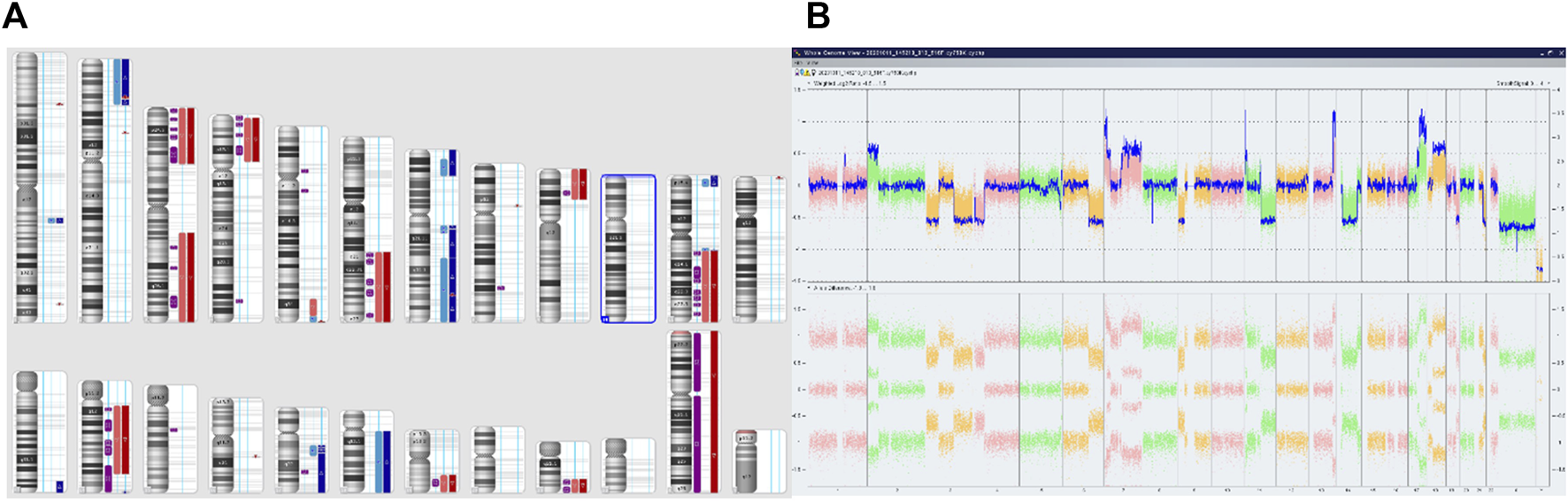
Complex copy number gains and losses in the whole genome of patient 64 with Neuroblastoma. Colored segmental regions (top line: red color—deletions; blue color—gains) (A) and Whole Genome View (B).
TABLE 2
| Molecular karyotype | ||
|---|---|---|
| Different aberrations | Common aberrations | |
| First cell line | arr [GRCh37] 1q21.3q23.1 (153001296_157812399)x3 [0.7], 2p25.3p21 (12771_42229545)x3 [0.7], 3p26.3p21.2 (61892_51747702)x1 [0.7], 3q13.31q29 (115403680_197851444)x1 [0.7], 4p16.3p13 (2744163_43086625)x1 [0.7], 6q21q27 (106487938_170914297)x1 [0.7], 7p21.3p15.3 (8582953_24331799)x3 [0.7], 7q11.22q36.3 (70144228_159119707)x3 [0.7], 9p24.3p21.1 (208455_28198623)x1 [0.7], 11p15.4 (3189610_10187920)x3 [0.7] 11q13.3q25 (69472473_134937416)x1 [0.7] 14q11.2q31.3 (24238818_89633472)x1 [0.7], 17q12q21.32 (36152227_46070727)x3 [0.7], 18q11.2q23 (19414307_78013728)x3 [0.7], 19q13.2q13.43 (42791601_58956816)x1 [0.7], 21q22.11q22.3 (35184499_48093361)x1 [0.7] |
arr [GRCh37] (Y)x0, 7p22.3p21.3 (43377_8573211)x3∼4, 11p15.5p15.4 (230681_3021529)x3∼4, 13q33.1q34 (103826264_115107733)x3∼4 14q31.1q32.33 (81449690_107279475)x2hmz 17q21.32q25.3 (46073099_81041823)x3∼4 |
| Second cell line | arr [GRCh37]11q13.2q13.3 (66814096_69464793)x3 [0.4] | |
| Third cell line | arr [GRCh37]5q33.3q35.2 (159230740_174855411)x1 [0.25], 5q35.2q35.3 (174856439_180715096)x3 [0.25] | |
Molecular changes identified in the microarray analysis of patient 64.
The third patient was a girl who was over 18 months of age at diagnosis, with stage 3 disease and tumor localization in the abdomen. Through fluorescence in situ hybridization we did not observe any amplification of the MYCN gene. Microarray analysis revealed mosaic monosomy of chromosome X and duplication of region 22q11.21 (arr [GRCh37] 22q11.21 (18979347_21800471)x3). The Whole Genome View (WGV) showed a decrease in the log2ratio value, which may indicate the presence of monosomy on chromosomes 3, 4, 9, 10, 14, 15, 19, and 21, and an increase in the log2ratio value, which may indicate the presence of trisomy on chromosome 13. An ambiguous assessment of chromosome copy number may be the result of incorrect material collection. The fourth patient was a boy who was over 18 months of age at diagnosis, with stage 3 disease and tumor localization in the abdomen. MYCN amplification was negative. With microarray analysis we identified a duplication involving a fragment of the short arm of chromosome 1 (arr [GRCh37]1p36.33p36.32 (849467_3627839)x3 [0.3]) and deletions in the following regions: arr [GRCh37]12q24.23q24.31 (118233114_125556278)x1 [0.2], 19p13.3p11 (260912_24505049)x1 [0.2]. As with the previous patient the Whole Genome View (WGV) showed a decrease in the log2ratio value, which may indicate the presence of monosomy on chromosomes 3, 4, 9, 10, 14, and 15 and an increase in the log2ratio value, which may indicate the presence of trisomy on chromosome 13. The microarray results for the two patients were normal and did not reveal any additional changes in the molecular karyotype.
Discussion
In recent years, significant progress has been made in understanding the genetic basis of neuroblastoma. Numerous studies have been published on somatic mutations and chromosomal abnormalities associated with the development of neuroblastoma. However, their role in treatment response, prognosis, and the development of metastasis has not been fully elucidated. Based on our research, we have divided patients into two groups: those with MYCN amplification and those without it.
High-risk patients are identified by the presence of MYCN amplification. MYCN amplification is a powerful prognostic factor for advanced stages and rapid tumor progression (Lau et al., 2015). Interphase FISH analysis provides a direct and rapid method. Only a small number of tumor cells are required, compared with primary tumor biopsies, and FISH is more sensitive by counting the MYCN copy number in each single cell (Tibiletti, 2007; Campbell et al., 2017).
The fluorescence in situ hybridization (FISH) method is faster to prepare than molecular biology techniques or traditional cytogenetic analysis. FISH can be used on interphase nuclei, in contrast to karyotype analysis, and it can be performed on formalin-fixed nuclei, in cases where a fresh or frozen tumor is not available (e.g., small biopsies or retrospective studies). However, molecular methods, such as microarrays, have strong limitations when analyzing MYCN amplification and other genetic changes in neuroblastoma because molecular methods are based on DNA from a mixture of cells and therefore only provide an average result for a particular tumor. When a tumor is highly amplified, this is not a problem for diagnosis. However, with low levels of MYCN amplification, it can be difficult to interpret the results. The first possibility is that there is a heterogeneous population of tumor cells in which a small number of cells are highly amplified or have other anomalies. Or it may be a highly amplified tumor mixed with normal tissue. Finally, it may be a low level of amplification occurring in tumor cells (e.g., a triploid population) (Shapiro et al., 1993; Squire et al., 1996; Thorner et al., 2006).
In the Polish population, tumor imprints are routinely used to assess MYCN amplification. However, this is not always possible and then the only material for MYCN amplification is a paraffin tissue section. It can be done on both fresh and archived tissues from many years ago.
For this study we used intraoperative tumor imprints and fresh paraffin-embedded tissue sections in addition to archival materials. Fresh material was obtained from a tumor removed from a patient 2 weeks before the study and archival paraffin sections were removed from a patient operated on 8 years and 2 months before the study. In our opinion both fresh tissue and tissue from years before preserved in a paraffin block were adequate materials for the test. The quality of the specimens was similar and the evaluation of both archival and fresh specimens was consistent with previous studies. Intraoperative tumor imprints were taken immediately after their removal. The quality of these specimens is much better compared to the paraffin-embedded tissue sections, as only the tumor cells are on the slide, with no surrounding tissue. The assessment of MYCN amplification on intraoperative tumor imprints does not cause any diagnostic problems.
Microarray testing of neuroblastoma samples provides additional information on other chromosomal changes observed in patients with and without MYCN amplification, which has a strong impact on prognosis. In a sample from a patient with MYCN amplification we found a deletion within the short arm of chromosome 1 (1p36.33p36.22), and a duplication involving a fragment of the short arm of chromosome 1 and the long arm of chromosome 1 (1p13.3q44). In the literature, deletion or loss of heterozygosity (LOH) of the 1p36 chromosomal band is correlated with high-risk neuroblastoma features, including older age at diagnosis, metastatic disease, MYCN amplification and poor prognosis Unfavorable abnormalities also include segmental copy number alterations, such as loss of 3p, deletion or LOH 11q23, 14q23qter and gain of 17q (Marrano et al., 2017). Amplification of the MYCN gene was observed only at a level of 10 copies. Unfortunately, this can be considered a limitation of the molecular method because the FISH analysis showed the amplification of the MYCN gene at a level of approximately 30 copies. In our opinion, the gold standard for the assessment of MYCN amplification is fluorescence in situ hybridization on intraoperative tumor imprints. Furthermore, microarray analysis showed a duplication of the 2p23.2p23.1 region, including the ALK gene. Anaplastic lymphoma kinase (ALK) is an important regulator of stem cell functions, including STAT3 dependent self-renewal, and as a transcriptional target of MYCN, high expression indicates an unfavorable prognosis (Rosswog et al., 2023). According to studies carried out on genetically engineered mouse models of NB, the MYCN and ALK genes cooperate to promote oncogenesis (Vivancos Stalin et al., 2019). Additionally, CytoScan HD analysis revealed a monosomy of chromosome 3 and a trisomy of chromosome 18.
In 3 out of 5 patients without changes in MYCN amplifications, we observed complex alterations in the microarray pattern. In two samples, discrete changes indicating the presence of monosomy of chromosomes 3, 4, 9, 10, 14, and 15. According to scientific research chromosome 10 is the most frequent loss. On the long arm of chromosome 10, there are tumor suppressor genes such as PTEN, DBMT, and LGI1, while the short arm contains IDI1, AKR1C3, DDH1, NET1A, PRKCQ, and GATA-binding protein 3. Among these, PTEN is the second most commonly deleted/mutated gene in human tumors, with a mutation rate of 50%. The GDNF family receptor alpha 2 (GFRA2) promotes the proliferation of NB cells (Li et al., 2019). According to the literature, patients with neuroblastoma and an abnormal chromosome 10 had a significantly lower 3-year OS compared to the group of patients with a normal copy number of chromosome 10 (Jiang et al., 2021). In the same samples we noted an extra copy of chromosome 13, which, according to other researchers (Bilke et al., 2008; Ambros et al., 2014) is a common chromosome in the trisomy of neuroblastoma. Interestingly, some authors show that there are no structural changes on chromosome 13 in neuroblastoma (Bogen et al., 2016). While its prognostic significance in neuroblastoma has not been precisely determined, in other cancers, including ALL, AML, and MDS, trisomy 13 is closely associated with an unfavorable prognosis for the patient. The presence of trisomy 13 in acute leukemia cells indicates a poor response to chemotherapy (Spirito et al., 2003; Herold et al., 2014). Monosomy of chromosome X and duplication of region 22q11.21 have not been described in the literature in patients with neuroblastoma so far. However, our results unequivocally demonstrate that such alterations may also be present in this type of cancer.
According to the Polish “Standards for the genetic diagnostics of solid tumors in children” structural changes involving chromosomal regions 1p, 1q, 2p, 3p, 4p, 6p, 6q, 11q, and 17q are most commonly observed in advanced stages of the disease in older children and represent unfavorable prognostic factors, increasing the risk of disease recurrence, regardless of the specific chromosomal pairs affected (Trubicka et al., 2023). In one of our patients with stage 4 disease, the FISH study found no amplification of the MYCN gene, while the microarray analysis identified 3 cell lines in which some of the above-mentioned structural changes were noted (including: 1p, 2p, 3p, 4p, 6q, 11q, and 17q) stratifying the patient into a group with an unfavorable prognosis.
In the literature, high-risk patients had long-term survival rates of only 10%–20% with combination chemotherapy, surgery, and local radiation therapy (Tolbert and Matthay, 2018). However, treatment approaches that used a combination of induction therapy, myeloablative consolidation therapy with stem cell support, and biological therapy had improved 5-year survival rates from less than 15%–40% (Nakagawara et al., 2018).
Despite many biochemical, molecular and genetic indicators as markers of poor prognosis, MYCN amplification is still used to stratify patients into risk groups and is a marker of poor prognosis. These patients respond poorly to conventional therapy. The ability to determine this biological marker is necessary for appropriate clinical management. Unfortunately, treatment failure has been reported in all patient groups, even without MYCN amplification, suggesting that additional prognostic markers must be discovered to improve treatment protocols (Feng et al., 2023).
In conclusion, both the FISH technique and microarray testing have their advantages and disadvantages. In addition to MYCN gene amplification, cytogenetic abnormalities are also a marker of an unfavorable prognosis in patients diagnosed with neuroblastoma. Therefore, it is very important that the result be based on both, fluorescence in situ hybridization and molecular karyotype.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the Medical University of Lublin (KE-0254/229/2012). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
DW and ML are responsible for the conception and design of the study. DW and BS conducted laboratory work. BS and PS were responsible for the acquisition of literatures for manuscript. DW and BS were responsible for interpretation of data. DW, PS, MS, and ML prepared final manuscript for publication. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
Preprint: “Winnicka et al., 2020” is part of this article, which has been enriched with additional research and updated.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/abp.2024.12821/full#supplementary-material
References
1
Adamkiewicz – Drożyńska E. (2006). Ocena znaczenia wybranych wskaźników proliferacji i angiogenezy w guzach z grupy nerwiaka zarodkowego współczulnego dla prognozowania klinicznego. Ann. Acad. Med. Gedanensis5, 1–208.
2
Ahmed A. A. Zhang L. Reddivalla N. Hetherington M. (2017). Neuroblastoma in children: update on clinicopathologic and genetic prognostic factors. Pediatr. Hematol. Oncol.34 (3), 165–185. 10.1080/08880018.2017.1330375
3
Ambros I. M. Brunner C. Abbasi R. Frech C. Ambros P. F. (2014). Ultra-high density SNParray in neuroblastoma molecular diagnostics. Front. Oncol.4, 202. 10.3389/fonc.2014.00202
4
Ambros P. F. Ambros I. M. Brodeur G. M. Haber M. Khan J. Nakagawara A. et al (2009). International consensus for neuroblastoma molecular diagnostics: report from the international neuroblastoma risk group (INRG) biology committee. Br. J. Cancer100 (9), 1471–1482. 10.1038/sj.bjc.6605014
5
Aygun N. (2018). Biological and genetic features of neuroblastoma and their clinical importance. Curr. Pediatr. Rev.14 (2), 73–90. 10.2174/1573396314666180129101627
6
Bhat N. McGregor L. (2017). Neuroblastoma: the clinical aspects. Neuroblastoma—current state and recent updates. InTech. 10.5772/intechopen.70486
7
Bilke S. Chen Q. R. Wei J. S. Khan J. (2008). Whole chromosome alterations predict survival in high-risk neuroblastoma without MYCN amplification. Clin. Cancer Res.14 (17), 5540–5547. 10.1158/1078-0432.CCR-07-4461
8
Bogen D. Brunner C. Walder D. Ziegler A. Abbasi R. Ladenstein R. L. et al (2016). The genetic tumor background is an important determinant for heterogeneous MYCN-amplified neuroblastoma. Int. J. cancer139 (1), 153–163. 10.1002/ijc.30050
9
Brodeur G. M. Seeger R. C. Schwab M. Varmus H. E. Bishop J. M. (1985). Amplification of N-myc sequences in primary human neuroblastomas: Correlation with advanced disease stage. Prog. Clin. Biol. Res.175, 105–113.
10
Brunner C. Brunner-Herglotz B. Ziegler A. Frech C. Amann G. Ladenstein R. et al (2016). Tumor touch imprints as source for whole genome analysis of neuroblastoma tumors. PloS one11 (8), e0161369. 10.1371/journal.pone.0161369
11
Campbell K. Gastier-Foster J. M. Mann M. Naranjo A. H. Van Ryn C. Bagatell R. et al (2017). Association of MYCN copy number with clinical features, tumor biology, and outcomes in neuroblastoma: a report from the children's Oncology group. Cancer123 (21), 4224–4235. 10.1002/cncr.30873
12
Chicard M. Boyault S. Colmet Daage L. Richer W. Gentien D. Pierron G. et al (2016). Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin. Cancer Res.22 (22), 5564–5573. 10.1158/1078-0432.CCR-16-0500
13
Croteau N. Nuchtern J. LaQuaglia M. P. (2021). Management of neuroblastoma in pediatric patients. Surg. Oncol. Clin. N. Am.30 (2), 291–304. 10.1016/j.soc.2020.11.010
14
Feng L. Li S. Wang C. Yang J. (2023). Current status and future perspective on molecular imaging and treatment of neuroblastoma. Seminars Nucl. Med.53 (4), 517–529. 10.1053/j.semnuclmed.2022.12.004
15
Herold T. Metzeler K. H. Vosberg S. Hartmann L. Röllig C. Stölzel F. et al (2014). Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood124 (8), 1304–1311. 10.1182/blood-2013-12-540716
16
Huang M. Weiss W. A. (2013). Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med.3 (10), a014415. 10.1101/cshperspect.a014415
17
Jeison M. Ash S. Halevy-Berko G. Mardoukh J. Luria D. Avigad S. et al (2010). 2p24 gain region harboring MYCN gene compared with MYCN amplified and nonamplified neuroblastoma: biological and clinical characteristics. Am. J. Pathology176 (6), 2616–2625. 10.2353/ajpath.2010.090624
18
Jiang Cy. Xu X. Jian Bl. Zhang X. Yue Z. X. Guo W. et al (2021). Chromosome 10 abnormality predicts prognosis of neuroblastoma patients with bone marrow metastasis. Italian J. Pediatr.47 (1), 134. 10.1186/s13052-021-01085-6
19
Kaczówka P. Wieczorek A. Czogała M. Książek T. Szewczyk K. Balwierz W. (2018). The role of N-Myc gene amplification in neuroblastoma childhood tumour - single-centre experience. Współczesna Onkol.22 (4), 223–228. 10.5114/wo.2018.81402
20
Lau D. T. Flemming C. L. Gherardi S. Perini G. Oberthuer A. Fischer M. et al (2015). MYCN amplification confers enhanced folate dependence and methotrexate sensitivity in neuroblastoma. Oncotarget6 (17), 15510–15523. 10.18632/oncotarget.3732
21
Lee J. W. Son M. H. Cho H. W. Ma Y. E. Yoo K. H. Sung K. W. et al (2018). Clinical significance of MYCN amplification in patients with high-risk neuroblastoma. Pediatr. blood cancer65 (10), e27257. 10.1002/pbc.27257
22
Lejman M. Włodarczyk M. Styka B. Pastorczak A. Zawitkowska J. Taha J. et al (2020). Advantages and limitations of SNP array in the molecular characterization of pediatric T-cell acute lymphoblastic leukemia. Front. Oncol.10, 1184. 10.3389/fonc.2020.01184
23
Lerone M. Ognibene M. Pezzolo A. Martucciello G. Zara F. Morini M. et al (2021). Molecular genetics in neuroblastoma prognosis. Child. (Basel, Switz.)8 (6), 456. 10.3390/children8060456
24
Li Z. Xie J. Fei Y. Gao P. Xie Q. Gao W. et al (2019). GDNF family receptor alpha 2 promotes neuroblastoma cell proliferation by interacting with PTEN. Biochem. biophysical Res. Commun.510 (3), 339–344. 10.1016/j.bbrc.2018.12.169
25
Lim H. Son M. H. Hyun J. K. Cho H. W. Ju H. Y. Lee J. W. et al (2020). Clinical significance of segmental chromosomal aberrations in patients with neuroblastoma: first report in Korean population. J. Korean Med. Sci.35 (14), e82. 10.3346/jkms.2020.35.e82
26
Marrano P. Irwin M. S. Thorner P. S. (2017). Heterogeneity of MYCN amplification in neuroblastoma at diagnosis, treatment, relapse, and metastasis. Genes, chromosomes cancer56 (1), 28–41. 10.1002/gcc.22398
27
Morandi F. (2022). Molecular determinants of neuroblastoma. Int. J. Mol. Sci.23 (7), 3751. 10.3390/ijms23073751
28
Nakagawara A. Li Y. Izumi H. Muramori K. Inada H. Nishi M. (2018). Neuroblastoma. Jpn. J. Clin. Oncol.48 (3), 214–241. 10.1093/jjco/hyx176
29
Otte J. Dyberg C. Pepich A. Johnsen J. I. (2020). MYCN function in neuroblastoma development. Front. Oncol.10, 624079. 10.3389/fonc.2020.624079
30
Park J. R. Bagatell R. Cohn S. L. Pearson A. D. Villablanca J. G. Berthold F. et al (2017). Revisions to the international neuroblastoma response criteria: a consensus statement from the national cancer Institute clinical trials planning meeting. J. Clin. Oncol.35 (22), 2580–2587. 10.1200/JCO.2016.72.0177
31
Rosswog C. Fassunke J. Ernst A. Schömig-Markiefka B. Merkelbach-Bruse S. Bartenhagen C. et al (2023). Genomic ALK alterations in primary and relapsed neuroblastoma. Br. J. cancer128 (8), 1559–1571. 10.1038/s41416-023-02208-y
32
Schulte M. Köster J. Rahmann S. Schramm A. (2018). Cancer evolution, mutations, and clonal selection in relapse neuroblastoma. Cell Tissue Res.372, 263–268. 10.1007/s00441-018-2810-5
33
Shapiro D. N. Valentine M. B. Rowe S. T. Sinclair A. E. Sublett J. E. Roberts W. M. et al (1993). Detection of N-myc gene amplification by fluorescence in situ hybridization. Diagnostic utility for neuroblastoma. Am. J. pathology142 (5), 1339–1346.
34
Spirito F. R. Mancini M. Derme V. Cimino G. Testi A. M. Tafuri A. et al (2003). Trisomy 13 in a patient with common acute lymphoblastic leukemia: Description of a case and review of the literature. Cancer Genet. Cytogenet.144 (1), 69–72. 10.1016/s0165-4608(02)00924-x
35
Squire J. A. Thorner P. Marrano P. Parkinson D. Ng Y. K. Gerrie B. et al (1996). Identification of MYCN copy number heterogeneity by direct FISH analysis of neuroblastoma preparations. Mol. Diagn.1 (4), 281–289. 10.1016/s1084-8592(96)70010-3
36
Swift C. C. Eklund M. J. Kraveka J. M. Alazraki A. L. (2018). Updates in diagnosis, management, and treatment of neuroblastoma. RadioGraphics38 (2), 566–580. 10.1148/rg.2018170132
37
Thorner P. S. Ho M. Chilton-MacNeill S. Zielenska M. (2006). Use of chromogenic in situ hybridization to identify MYCN gene copy number in neuroblastoma using routine tissue sections. Am. J. Surg. Pathology30 (5), 635–642. 10.1097/01.pas.0000202163.82525.5c
38
Tibiletti M. G. (2007). Interphase FISH as a new tool in tumor pathology. Cytogenet. genome Res.118 (2-4), 229–236. 10.1159/000108305
39
Tolbert V. P. Matthay K. K. (2018). Neuroblastoma: Clinical and biological approach to risk stratification and treatment. Cell tissue Res.372 (2), 195–209. 10.1007/s00441-018-2821-2
40
Trubicka J. Młynarski W. Lejman M. Balwierz W. Małdyk J. Pastorczak A. et al (2023). Standards for the genetic diagnostics of solid tumors in children. Pediatr. Rev.52 (3), 6–27.
41
Van Noesel M. M. Versteeg R. (2004). Pediatric neuroblastomas: genetic and epigenetic “danse macabre”. Gene325 (1-2), 1–15. 10.1016/j.gene.2003.09.042
42
Vivancos Stalin L. Gualandi M. Schulte J. H. Renella R. Shakhova O. Mühlethaler-Mottet A. (2019). Expression of the neuroblastoma-associated ALK-F1174L activating mutation during embryogenesis impairs the differentiation of neural crest progenitors in sympathetic ganglia. Front. Oncol.9, 275. 10.3389/fonc.2019.00275
43
Whittle S. B. Smith V. Doherty E. Zhao S. McCarty S. Zage P. E. (2017). Overview and recent advances in the treatment of neuroblastoma. Expert Rev. anticancer Ther.17 (4), 369–386. 10.1080/14737140.2017.1285230
44
Winnicka D. Babicz M. Styka B. Skowera P. Zawitkowska J. Lejman M. (2020). Application of fluorescence in situ hybridization to detect MYCN amplification on paraffin-embedded tissue sections of neuroblastoma - single Institute study. Research Square [Preprint]. Version 1. 10.21203/rs.3.rs-95672/v1 (accessed October 26, 2020).
45
Yue Z. X. Huang C. Gao C. Xing T. Y. Liu S. G. Li X. J. et al (2017). MYCN amplification predicts poor prognosis based on interphase fluorescence in situ hybridization analysis of bone marrow cells in bone marrow metastases of neuroblastoma. Cancer Cell Int.17, 43. 10.1186/s12935-017-0412-z
Summary
Keywords
neuroblastoma, childhood tumours, MYCN amplification, high-density SNP arrays, FISH
Citation
Winnicka D, Skowera P, Stelmach M, Styka B and Lejman M (2024) Application of the FISH method and high-density SNP arrays to assess genetic changes in neuroblastoma—research by one institute. Acta Biochim. Pol 71:12821. doi: 10.3389/abp.2024.12821
Received
08 February 2024
Accepted
12 June 2024
Published
10 July 2024
Volume
71 - 2024
Edited by
Witold Norbert Nowak, Jagiellonian University, Poland
Reviewed by
Walentyna Balwierz, University Children’s Hospital in Krakow, Poland
Artur Palasz, Medical University of Silesia, Poland
Hevidar Taha, University of Duhok, Iraq
Updates
Copyright
© 2024 Winnicka, Skowera, Stelmach, Styka and Lejman.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paulina Skowera, paulina.skowera@umlub.pl; Magdalena Stelmach, magdalena.stelmach@umlub.pl
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.