Abstract
Background:
Duck enteritis virus (DEV) belongs to Alphaherpesvirinae; little is known about the DEV UL7 gene and its encoded protein. This study examined the molecular characteristics of DEV pUL7 in vitro and determined whether DEV pUL7 co-localizes with pUL51.
Results:
The results showed that UL7 can be regarded as a late gene. Moreover, immunofluorescence assay revealed that pUL7 was located around the perinuclear cytoplasmic region and co-localized with pUL51 in the cytoplasm and nucleus after transfection into duck embryo fibroblast cells (DEFs).
Conclusion:
In conclusion, we identified the molecular characteristics of the DEV UL7 gene, which is a late gene, and the co-localization of its encoded protein with pUL51 in transfected DEFs, enriching our understanding of pUL7 and future research directions.
Introduction
Duck viral enteritis (DVE), also known as duck plague (DP), is an acute contagious infection of the order Anseriformes (ducks, geese, and swans) caused by Anatid herpesvirus 1 (DEV), a member of the subfamily Alphaherpesvirinae and Mardivirus genus (Wu et al., 2012; Dhama et al., 2017). The DEV genome is a linear double-stranded DNA that contains unique long and short (UL and US, respectively) regions, with each unique sequence bracketed by internal and terminal inverted repeat sequences (IRS and TRS, respectively): UL–IRS–US–TRS (Wu et al., 2012; Liu et al., 2015). The DEV strain CHv genome encodes 78 open reading frames (ORFs) (Wu et al., 2012) and many DEV genes and their potential proteins have been characterized (Deng et al., 2020; Liu et al., 2020; Shen et al., 2021; Yang et al., 2021; Zhou et al., 2021; He et al., 2022; Ning et al., 2022; Wan et al., 2022). However, the DEV UL7 protein (pUL7) molecule has not been characterized yet.
The UL7 gene encodes a 33 kDa tegument protein expressed late during infection and is conserved in all herpesviruses (Nozawa et al., 2002; Tanaka et al., 2008; Xingli Xu et al., 2016; Albecka et al., 2017). pUL7 is involved in the maturation and egress of the pseudorabies virus (PRV). Previous studies have demonstrated that herpes simplex virus 1 (HSV-1) pUL7 interacts with the mitochondrial adenine nucleotide transporter 2 (ANT2) using pull-down and co-immunoprecipitation assays (Fuchs et al., 2005; Tanaka et al., 2008). Herpes simplex virus 2 (HSV-2) pUL7 plays a supplementary role in viral DNA cleavage and packing (Nozawa et al., 2002). Bovine herpesvirus 1 (BHV-1) is a non-structural protein (Schmitt and Keil, 1996). The deduced amino acids sequence of DEV UL7 gene showed significant homology to that of HSV-1 UL7 gene with 34% identity (Plummer et al., 1998). Moreover, UL6 and UL7 share the same poly-A signal sequence located downstream of the UL7 gene (Li et al., 2009). HSV-1 pUL7 forms a complex with another tegument protein, the UL51 protein (pUL51) (Roller and Fetters, 2015; Albecka et al., 2017; Feutz et al., 2019; Butt et al., 2020), which can promote virus assembly by simulating the cytoplasmic wrapping of newly virions (Butt et al., 2020). In addition, the amino acids 30–90 of pUL51 mediate its interaction with pUL7 (Feutz et al., 2019). pUL7–pUL51 complex loss can be inhibited in secondary envelopment, which also identifies that the complex regulates focal adhesion stability during HSV-1 infection (Albecka et al., 2017). Thus, this complex plays an important role in the lifespan of HSV-1 cells.
The goal of this study was to describe the molecular characterization of DEV pUL7 in vitro and determine whether DEV pUL7 co-localized with pUL51, which will provide insight into further research of DEV pUL7.
Materials and methods
Cells and viruses
Duck embryo fibroblasts (DEFs) were cultured in minimum essential medium (MEM; Thermo Fisher Scientific, United States) supplemented with 10% newborn calf serum (NBS) at 37°C in a 5% CO2 incubator. Maintenance medium supplemented with 2% NBS was used for the viral infection. DEV strain CHv (GenBank: JQ647509.1) was isolated, preserved, and supplied by our laboratory.
Vectors and antibodies
The E. coli strain DH5α; E.coli BL21pLysS, pMD20-T, pET-32a (+), and pCAGGS vectors; and pCAGGS–UL51–Flag plasmid were provided by our laboratory. A rabbit anti-UL7 polyclonal antibody was generated for this study; rabbit anti-DEV and anti-UL51 polyclonal antibodies were prepared as described earlier (Shen et al., 2009). HRP-conjugated goat anti-rabbit IgG (Life Technologies, United States), mouse anti-β-actin (Beyotime, CHN), FITC-conjugated anti-rabbit IgG (Abcam, UK), anti-HA-tagged rabbit (Beyotime, CHN), anti-Flag-tagged mouse (Transgen Biotech, CHN), Alexa Fluor 488-conjugated goat anti-mouse IgG (Thermo Fisher Scientific, United States), and Alexa Fluor 594-conjugated goat anti-rabbit IgG (Life Technologies, United States) were used in this study.
Plasmids and transfection
The primers were designed with Oligo 7.0 and primer 5.0 (Table 1). The UL7 gene (GenBank: FJ222445) of the DEV strain CHv was inserted into the pMD20-T clone vector, digested with BamHI and XhoI, and ligated into E. coli expression vector pET-32a (+) (Invitrogen) to create the plasmid pET32a–UL7. The plasmids expressing UL7–HA and UL51–Flag from DEV were constructed by PCR amplification of the coding region and subcloned into the pCAGGS vector to create pCAGGS–HA–UL7 and pCAGGS–UL51–Flag, respectively, and transfected into DEFs using the HieffTrans™ liposomal transfection reagent (Yeasen, CHN) according to the manufacturer’s protocol.
TABLE 1
| Gene | Primer sequence (5′-3′) | Product size (bp) |
|---|---|---|
| Duck β-actin | CCGGGCATCGCTGACA | 177 |
| GGATTCATCATACTCCTGCTTGCT | ||
| DEV UL54 | GAACAACCGCCGAACAC | 127 |
| TCAAACATCCGCCTCAA | ||
| DEV UL13 | GCCACCAACCCTACCAAG | 131 |
| GTCGTCAGCCCATCACCA | ||
| DEV US2 | AGACGGTTCCGAAAGTACAG | 111 |
| TCGGCAGCACCAATAATCC | ||
| DEV UL7 | GGGACAAACCATCCTCTG | 114 |
| TATCGCCTGCCAACTTAT |
Oligonucleotide primers in this study.
Prokaryotic expression of pUL7
The recombinant pUL7 of DEV was purified under denaturing conditions because it was expressed in E. coli BL21pLysS with inclusion bodies. To obtain the highest pUL7 expression, the conditions were optimized by inducing different IPTG concentrations, temperatures, and time periods. All expression levels were detected by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (SDS–PAGE).
Polyclonal anti-UL7 sera preparation and identification
The DEV pUL7 was purified by gel and electric elution and used to prepare the polyclonal antiserum of pUL7. Briefly, four male New Zealand white rabbits (Chengdu dossy experimental animals CO., Ltd.) were immunized intradermally with 0.5 mg pUL7 emulsified in Freund’s complete adjuvant. Subsequently, Freund’s incomplete adjuvant and 1.0 mg pUL7 were injected. Two boosted subcutaneous injections were administered at 2 weeks interval after the primary injection. One week later, the rabbits were intravenously injected with 0.1 mg purified pUL7. Two weeks after the last immunization, the rabbits were bled and antiserum was prepared. Control preimmune serum samples were obtained from non-vaccinated healthy rabbits. The rabbit polyclonal antiserum was purified using ammonium sulfate precipitation (McGuire et al., 1996). In addition, the antibody titer of the pUL7 antiserum was confirmed using a double-diffusion assay in 1% agarose.
Western blotting
Protein samples were lysed in SDS sample buffer, separated by SDS–PAGE, and immediately transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, MA, United States). Nonspecific protein binding was blocked by treating the membranes with PBS containing 5% skim milk at room temperature for 1 h. The membranes were washed with PBST (PBS containing 0.05% Tween 20) and incubated overnight at 4°C with appropriately diluted primary antibodies. After washing three times with PBST, the membrane was incubated with the HRP-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibodies at 37°C for 1 h. The membranes were then washed three times with PBST, treated with an enhanced chemiluminescence (ECL) kit, and exposed on a BioRad CheminDOC XRS+ system.
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated from DEV-infected cells at 0, 1, 2, 4, 8, 12, 16, 24, 32, 40, 48, 56, and 64 h post-infection using a Total RNA Isolation System (Takara, JPN). The purified RNA was immediately inverse transcribed to cDNA by PrimeScript® RT reagent kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. The primers were designed by Oligo 7 (Table 1), and RT-qPCR was performed in 20 μL reaction volumes containing 1 μL each primer, 1 μL cDNA, 10 μL SYBR Green Mix, and 7 μL RNase-free water in triplicate. β-actin was used as an internal control. Average relative content of DEV UL7 gene transcription was calculated by the 2−ΔΔCT method (Liu et al., 2015).
Drug treatment
To examine the DEV UL7 gene expression patterns, DEFs were infected with the DEV strain CHv in three T25 flasks; one flask was treated with 300 μg/mL acyclovir (ACV), the second was treated with 100 μg/mL cycloheximide (CHX), while the third was prepared without any drugs as control. Total RNA was extracted from DEV-infected DEFs and the control group at 24 h post-infection (hpi) and subsequently reverse-transcribed into cDNA. PCR was performed using the primers for UL54, UL13, US2, UL7, and β-actin genes (Table 1).
Immunofluorescence microscopy
The cells were fixed overnight in 4% paraformaldehyde at 4°C after washing three times with PBS. The cells were then permeabilized with 0.2% Triton X-100 for 10 min and blocked with 3% bovine serum albumin (BSA) in PBS for 1 h at 37°C. The cells were labeled with primary antibodies at 4°C overnight and Alexa Fluor-conjugated secondary antibodies in blocking buffer for 1 h at 37°C. After that, the cells were stained with 4′6′-diamidino-2-phenylindole (DAPI) for 5 min after washing three times again at room temperature. Finally, the fluorescent images were examined using a Nikon H550L imaging system.
Immunoprecipitation
Proteins were immunoprecipitated from DEFs transfected with the pCAGGS–HA–UL7 and pCAGGS–UL51–Flag plasmids for 24 h. Transfected DEF monolayers of 50 mm cultures were washed twice with 3 mL cold PBST, and the cells were suspended in 1 mL cold PBST and pelleted at 110 × g for 10 min. The cell pellets were resuspended in 0.4 mL IP buffer (Yeasen, CHN) supplemented with PMSF (final concentration: 1 mM), transferred to new microcentrifuge tubes, and placed on ice for 5 min. Then, the cells were centrifuged at 10,000 × g for 10 min at 4°C, and the supernatant was transferred to a new tube. After removal of a fraction of the sample as a control, 2 µL mouse anti-Flag and rabbit anti-HA IgG were added and the mixture was incubated at 4°C overnight under gentle rotation. On the next day, 20 µL protein A agarose (Bio-Rad, United States) was added and the samples were incubated and gently rotated at 4°C for 3 h. The samples were then washed thrice with PBST, rapidly centrifuged for 30 s, and the supernatant was collected. Finally, the samples were heated for 10 min at 90°C with 5× SDS loading buffer.
Results
Rabbit anti-pUL7 antibody preparation
The recombinant plasmid pET32a–UL7 was transformed into E. coli BL21pLysS and expressed mainly in the insoluble fraction (50 kDa). pUL7 was purified and analyzed using SDS–PAGE (Figure 1A). The immunogenicity of the purified UL7 fusion protein was detected using rabbit anti-DEV antibodies, which specifically recognize a 50 kDa molecule (Figure 1B). Rabbit anti-pUL7 polyclonal antibodies (titer: 1:32) were generated in this study, which can specifically recognize pUL7 in DEV-infected DEFs (molecular weight: approximately 35 kDa), which is consistent with the expected size of DEV pUL7 (Figure 2D).
FIGURE 1
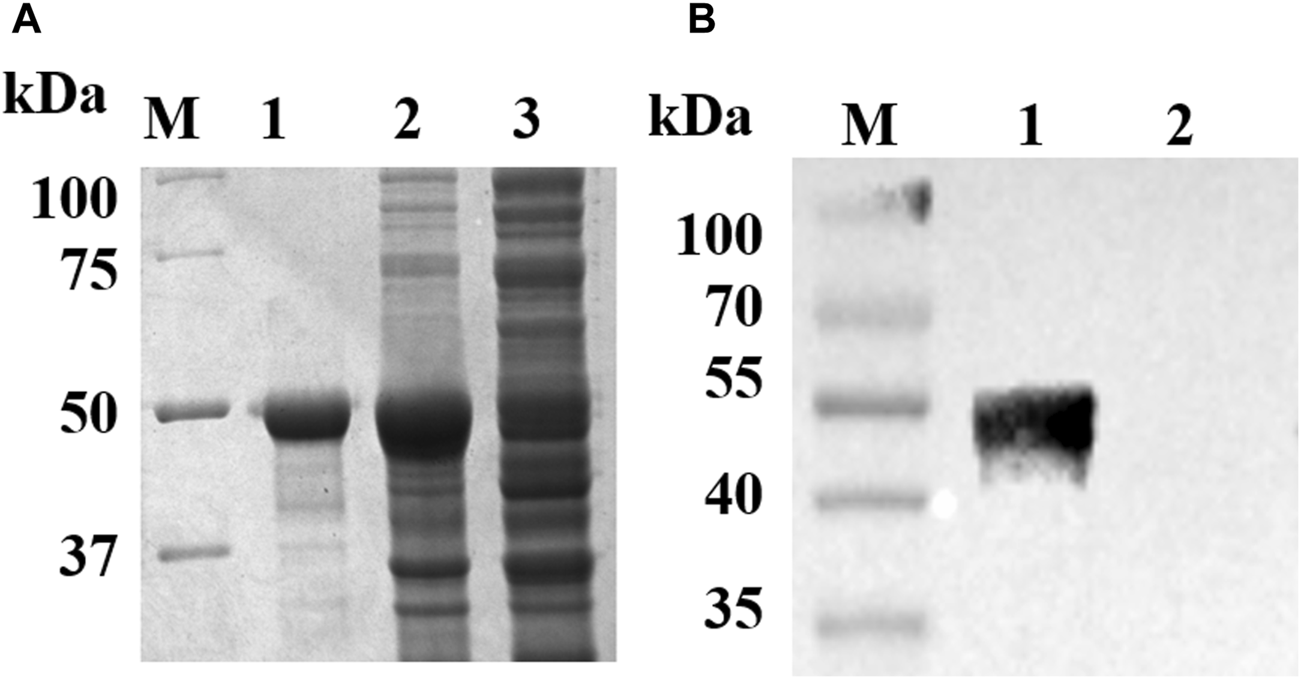
Identification of the recombinant pUL7. (A): Purification of the recombinant pUL7. M. Protein Marker; Lane 1, purified recombinant pUL7 (approximately 50 kDa); Lanes 2 and 3, the pellet and supernatant of expressed pUL7 after induction in E. coli BL21pLysS, respectively. (B): recombinant pUL7 identification by western blotting (rabbit anti-DEV antibody 1:800 and goat anti-rabbit IgG HRP-conjugated antibody 1:3000) M. Prestained protein Marker; Lane 1. Expression of recombinant pUL7 (approximately 50 kDa); Lane 2.pET32a (+) vector.
FIGURE 2
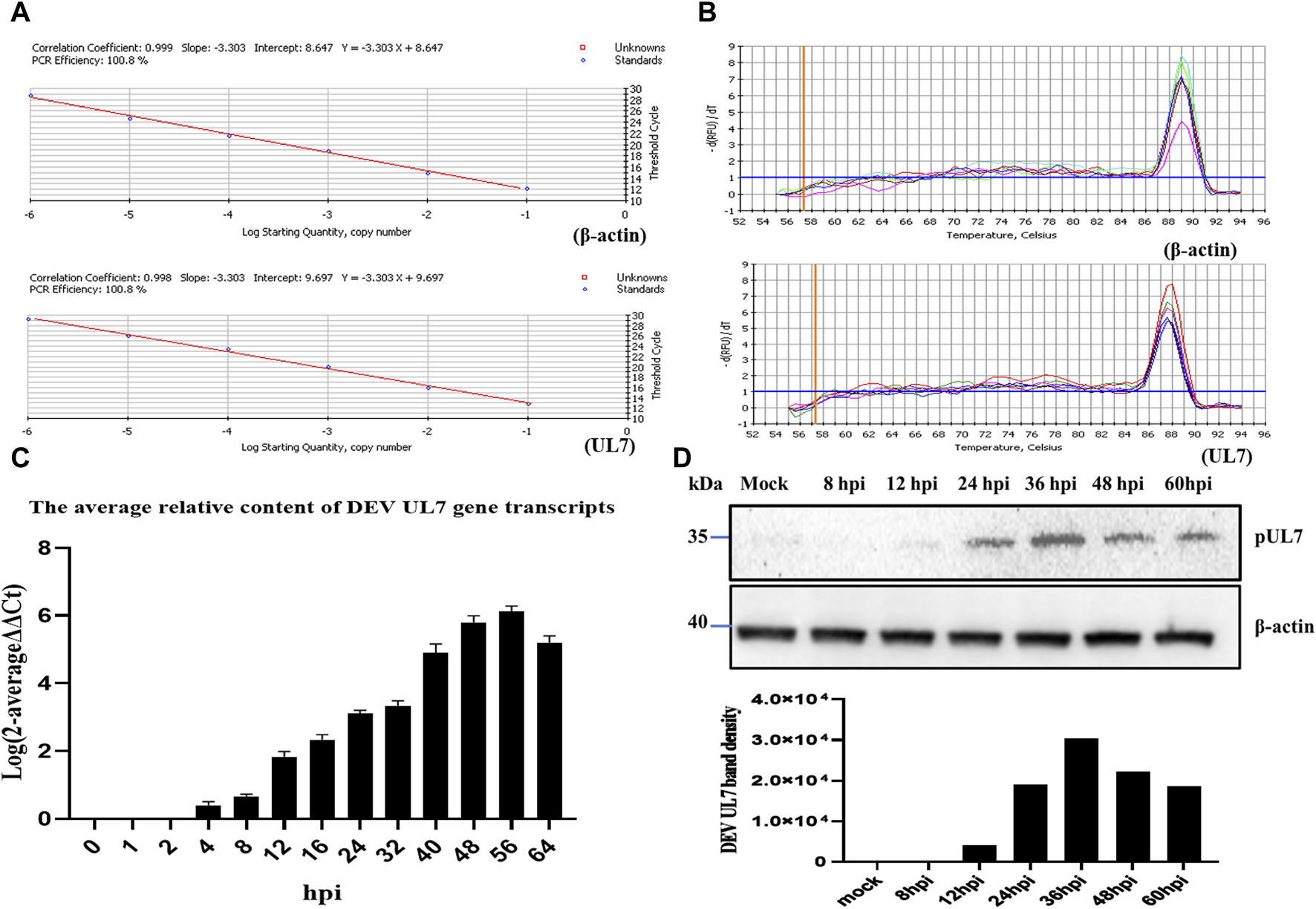
The transcription and expression kinetics of DEV UL7 gene. (A): Standard curves of DEV UL7 (Y = −3.303X + 9.697) and β-actin (Y = −3.303X + 8.647), and UL7 gene (100.8%) and β-actin (100.8%) amplification efficiencies were identical, with correlation coefficients of 0.998 and 0.999, respectively. (B): Melting curves of DEV UL7 gene and β-actin. (C): UL7 mRNA levels at 0, 1, 2, 4, 8, 12, 16, 24, 32, 40, 48, 56, and 64 phi were detected by RT-qPCR and normalized to β-actin, and average relative content of DEV UL7 gene transcription was calculated by the 2−ΔΔCt method. (D): The pUL7 express levels at 0, 8, 12, 24, 36, 48, and 60 phi were detected by western blotting (up, approximately 35 kDa) and quantification in the seven groups (down) were analyzed using Image J software. All cell lysate samples were detected with rabbit anti-UL7 serum (1:500) and goat anti-rabbit IgG HRP-conjugated antibody (1:3000). β-actin antibody (1:5000) was used to detect β-actin as a control.
Quantitative analysis of DEV UL7 gene transcripts
The amplification efficiency of the target and reference gene must be approximately equal for applying the 2−ΔΔCT method. Both standard curves showed the same PCR efficiency (Figure 2A) and their melting curves presented a single peak (Figure 2B). The UL7 transcripts in DEV-infected cells were analyzed by real-time quantitative PCR (RT-qPCR) with SYBR Green I. The average relative DEV UL7 gene transcript content was calculated by the 2−ΔΔCT method. The results indicated that DEV UL7 gene transcripts were not detected in negative controls (at 0 hpi), first detected at 4 hpi, increased steadily until reaching a peak at 56 hpi, and maintained a high level at 64 hpi with a descending trend (Figure 2C).
Kinetic class of DEV pUL7 expression levels
The cell lysates collected at different time points after DEV post-infection were analyzed by western blotting, and the PVDF membranes were probed with a rabbit anti-pUL7 polyclonal antibody (Figure 2D). Band density was analyzed using ImageJ software, which showed that pUL7 expression began at 12 hpi and peaked 36 hpi. As a control, the same cell lysates were probed with a monoclonal antibody against β-actin, and the β-actin levels remained relatively constant.
DEV UL7 is a late gene
Previous studies have shown that many DEV genes are late genes after acyclovir (ACV) or cycloheximide (CHX) treatment (He et al., 2018; Yang et al., 2020). Therefore, we examined the effects of ACV and CHX on DEV UL7. Figure 3 illustrates the correct bands of the immediate-early (IE) gene UL54, early (E) gene UL13, and late (L) gene US2; β-actin was detected as a control and the negative group was DEFs untreated with ACV or CHX. UL7 showed the same band as US2 and was observed only in the DEV-infected group without ACV or CHX treatment. These results indicated that UL7 is an L gene in DEV.
FIGURE 3
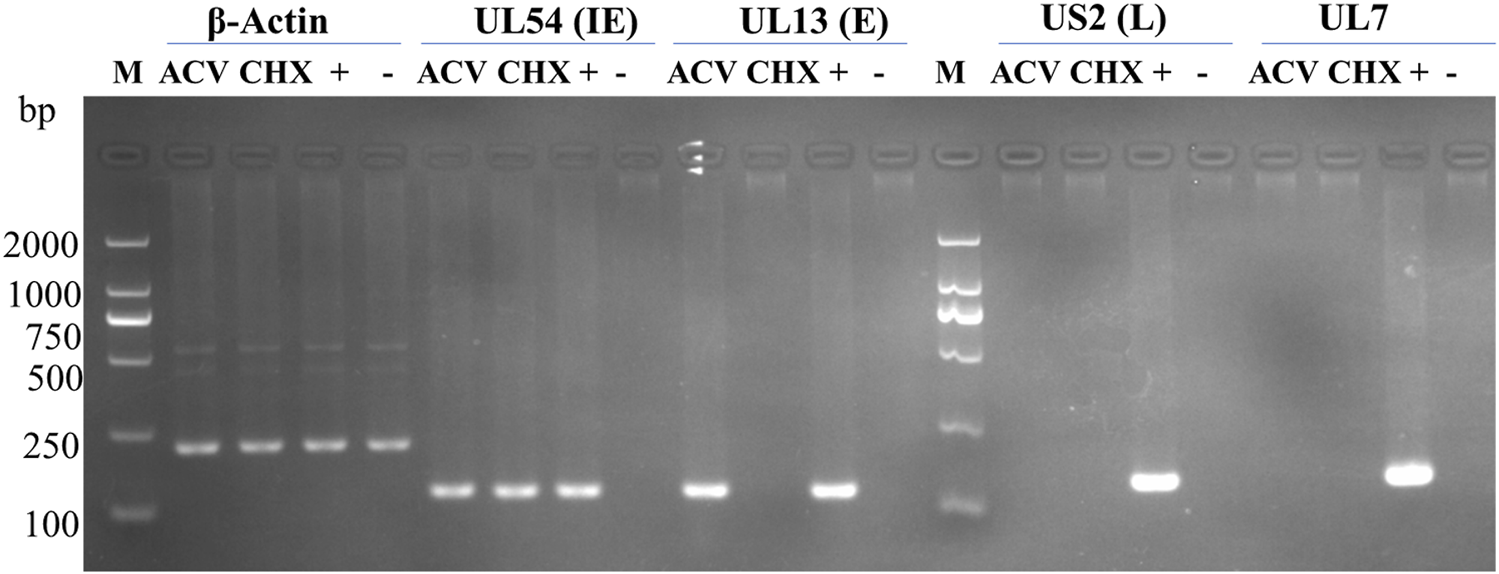
Identification virion gene type by drug treatment. M, DL2000 DNA Marker; ACV, DEV-infected cells treated with 300 μg/mL acyclovir (ACV); CHX, DEV- infected cells treated with 100 μg/mL cycloheximide (CHX). +, DEV-infected cells without drugs; −, untreated DEF cells. IE, immediate early; E, early; L, late.
pUL7 mainly localizes in the cytoplasm of DEV-infected cells
The intracellular localization of pUL7 in DEV-infected DEFs was examined by indirect immunofluorescence assays using pUL7 antiserum. The pUL7-specific fluorescent was detected in the cytoplasm at 8 hpi (Figure 4). At 48 hpi, the pUL7-specific fluorescent granules had increased and clustered strongly in the perinuclear region. However, no specific staining was observed in mock-infected DEFs and infected DEFs at 4 hpi. Taken together, these observations indicate that DEV pUL7 is predominantly localized in the cytoplasm of DEV-infected DEFs.
FIGURE 4
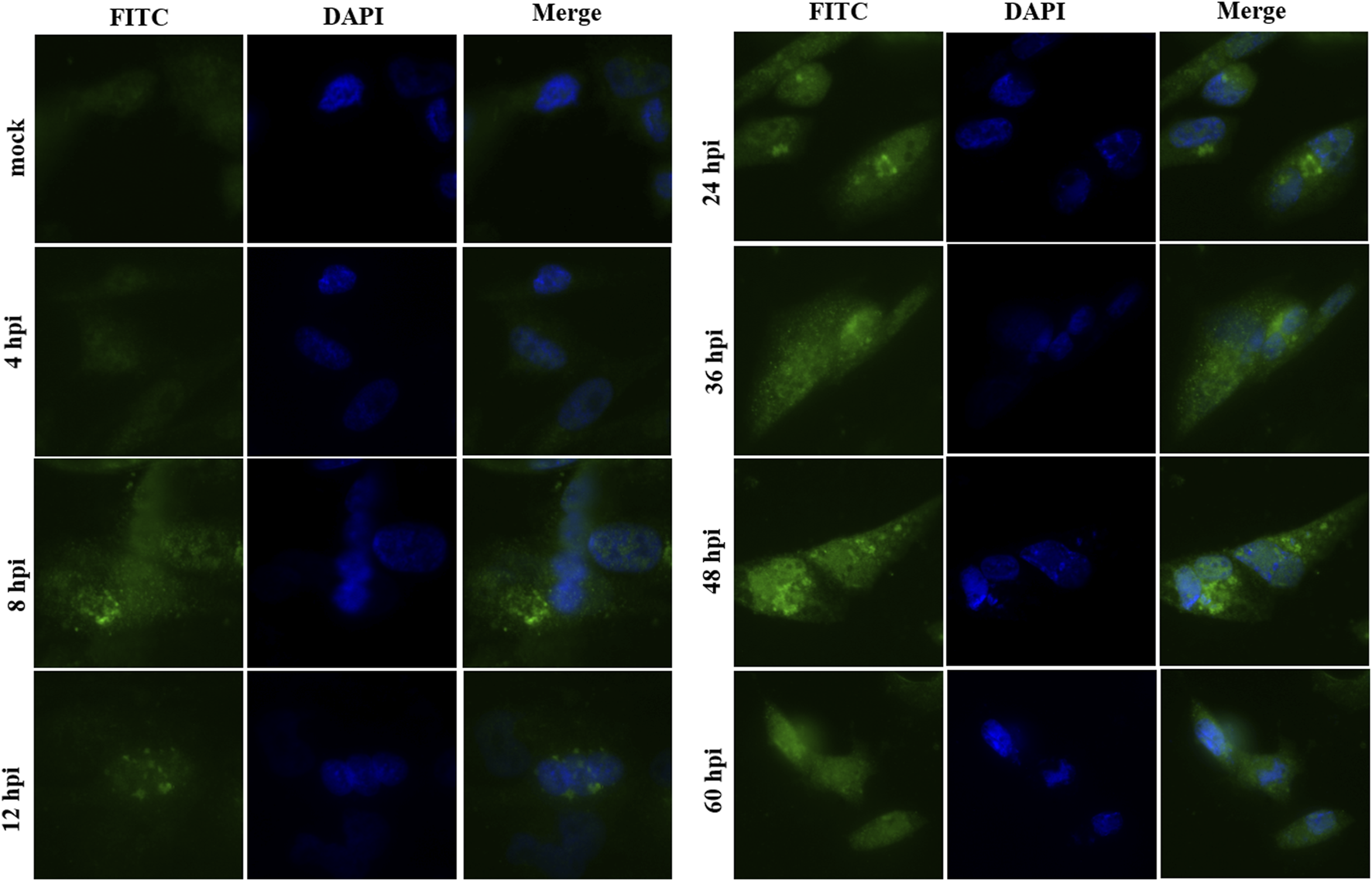
Intracellular localization of the pUL7 in DEV-infected DEFs at different times. Immunofluorescence microscopic images of the pUL7 (FITC green) at 4, 8, 12, 24, 36, 48, and 60 hpi. Nuclei (DAPI) are indicated in blue. The mock comprised untreated DEFs. The merged fluorescence microscopy images of DEFs were shown in panels with magnification (400×). All samples were detected with rabbit anti-UL7 antibody serum (1:100) and goat anti-rabbit IgG-conjugated FITC(1:1000). Fluorescent images were obtained with the Nikon H550L imaging system.
pUL7 co-localization with pUL51 in DEFs
To determine whether DEV pUL7 co-localization with pUL51, we first created pCAGGS–HA–UL7 and pCAGGS–UL51–Flag vectors and expressed them via transfection into DEFs individually or together. When cells were infected alone, pUL7 was mainly observed in the cytoplasm (Figure 4). In addition, pUL51 was detected mainly in the cytoplasm (Figure 5A). When pUL7 was co-expressed with pUL51, a juxtanuclear Golgi apparatus-like distribution was observed, and merged fluorescence analysis showed that the two proteins were co-localized in the cytoplasm and nucleus (Figure 5B). However, this did not necessarily imply an interaction during co-expression. To confirm that the pUL7–pUL51 complex could form in co-transfected DEFs, we detected the complex using co-immunoprecipitation (Co-IP). The results showed that pUL7 co-immunoprecipitated with pUL51 in the co-transfected DEFs (Figure 6).
FIGURE 5
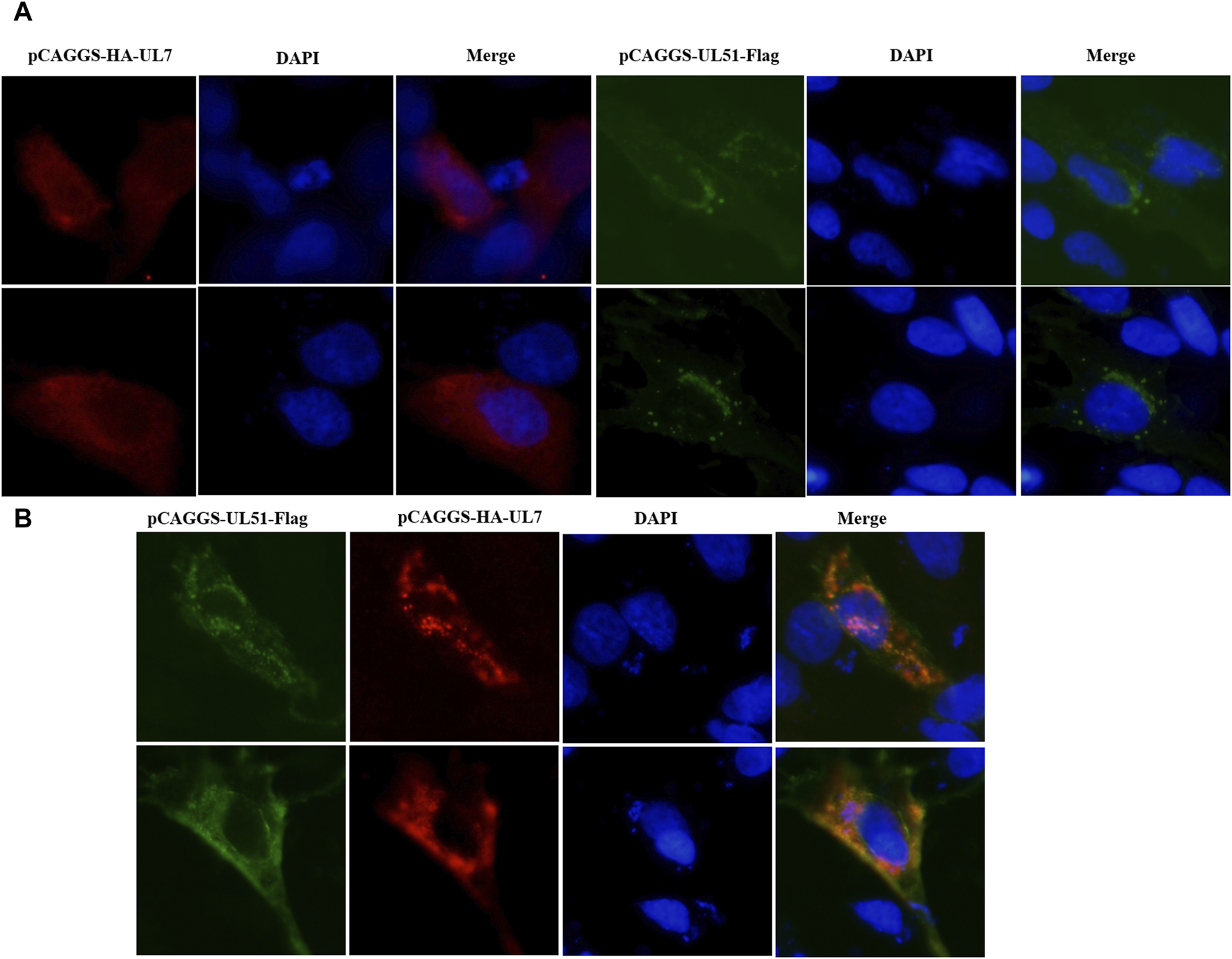
Colocalization of pUL7 with pUL51 in DEFs. (A): DEFs were transfected with pCAGGS-HA-UL7 and pCAGGS-UL51-Flag, respectively. (B): DEFs were co-transfected with pCAGGS-HA-UL7 and pCAGGS-UL51-Flag. All samples were collected after 24 h post transfection. Nuclei (DAPI) are indicated in blue. The merged fluorescence microscopy images of DEFs were shown in panels with magnification (400×). The pUL7 were detected with rabbit anti-HA antibody serum (1:500); pUL51 were detected with mouse anti-Flag antibody serum (1:500) and Alexa Fluor-conjugated secondary antibodies (1:1000). Fluorescent images were obtained with the Nikon H550L imaging system.
FIGURE 6
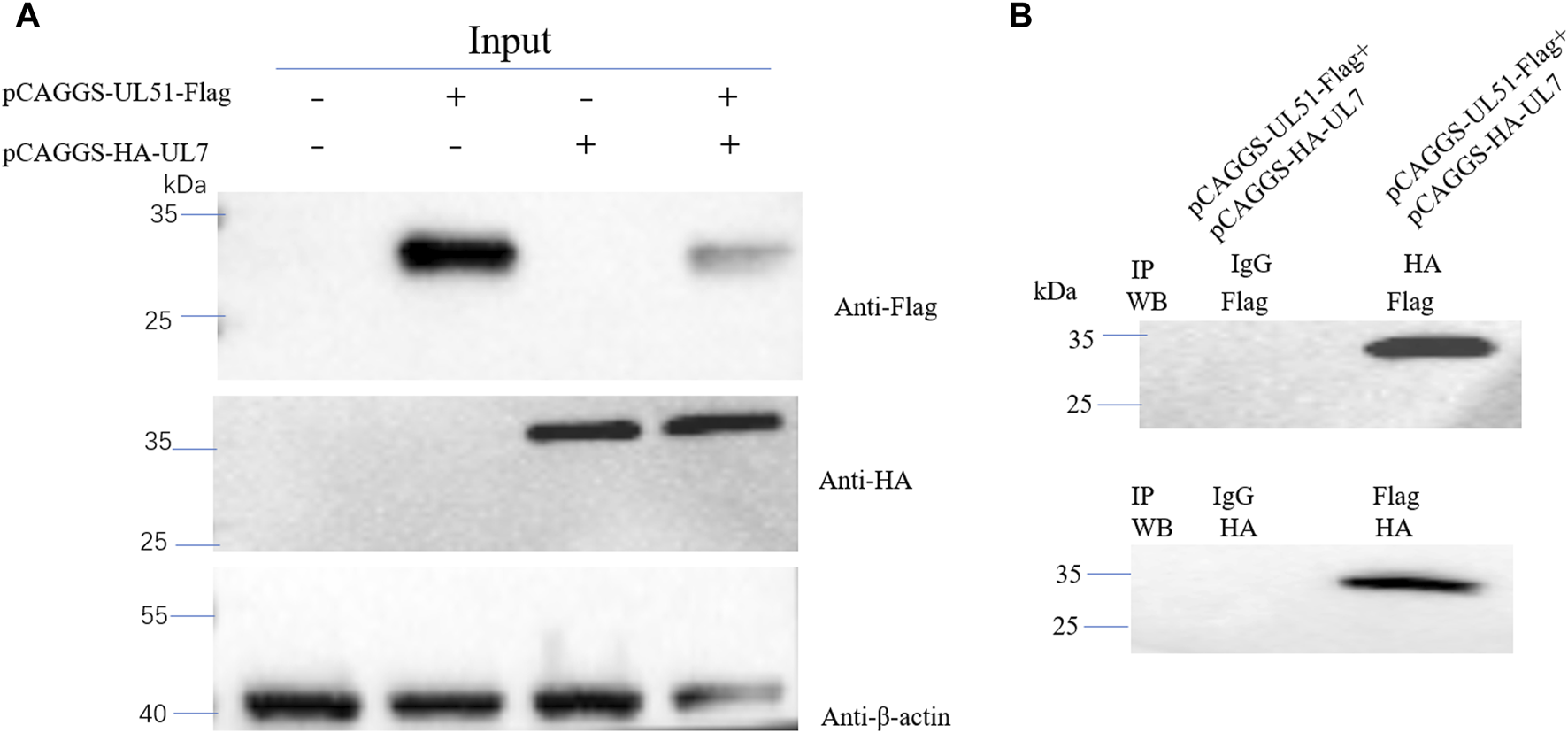
(A): DEFs were transfected or co-transfected with pCAGGS-UL51-Flag and pCAGGS-HA-UL7 expression plasmids. Anti-Flag, anti-HA and anti-β-actin monoclonal antibodies were used for WB. (B): DEFs were co-transfected with pCAGGS-UL51-Flag and pCAGGS-HA-UL7 expression plasmids. Anti-Flag and anti-HA monoclonal antibodies were used for IP, and anti-HA and anti-Flag monoclonal antibodies were used for WB, respectively.
Discussion
The UL7 gene is conserved in all herpesvirus, and the pUL7 or its homologous proteins in HSV-1, HSV-2, BHV-1, pseudorabies virus (PRV); ORF53 in varicella zoster virus (VZV); pUL103 in human cytomegalovirus (HCMV); and BBRF2 in Epstein–Barr virus (EBV) have been identified previously (Schmitt and Keil, 1996; Nozawa et al., 2002; Fuchs et al., 2005; Tanaka et al., 2008; Ahlqvist and Mocarski, 2011; Wang et al., 2017; Masud et al., 2019). A previous study demonstrated that the UL7 gene-null mutation is dispensable in HSV-1 replication in cells. However, both the plaque-forming ability and viral growth of the UL7-knockout HSV-1 were significantly impaired compared to those of the wild-type virus, which is consistent with the results of BHV-1 and PRV (Tanaka et al., 2008). The UL7 gene products of HSV-2, PRV, and HCMV are structural proteins. However, pUL7 was not found in the purified BHV-1 virions, although it accumulated predominantly in the cytoplasm, as detected by IFA (Schmitt and Keil, 1996). This difference may indicate the different roles of these proteins in different viruses. In this study, DEV pUL7 accumulated predominantly in the cytoplasm and may share a function with that of BHV-1.
Herpes virus genes are divided into three kinetic classes: IE, E, and L. The IE and E genes are involved in viral replication and expressed first. The L genes encoded structural proteins, which are subdivided into two categories: leaky-late (γ1) or strict-late (γ2). The γ1 genes can be expressed in the presence of viral DNA inhibitors. However, the γ2 genes cannot be expressed in the presence of viral DNA inhibitors (Lian et al., 2010; Gruffat et al., 2016). In general, viral DNA replication and protein synthesis inhibitors are used to distinguish between the gene classes. Viral DNA replication inhibitors include ACV, ganciclovir (GCV), and phosphonoacetic acid (PAA), but CHX is the only protein synthesis blocker. To distinguish between the viral E and L transcript stages, only the E transcripts were detected in cells infected in the presence of viral DNA replication inhibitors. Viral IE transcripts can be detected using CHX (Zhang et al., 2017). Several DEV genes, including US2 (Gao et al., 2015), UL16 (He et al., 2012), UL21 (Yang et al., 2020), UL41 (He et al., 2018), and UL55 (Wu et al., 2011), have been identified as L genes. In this study, we identified UL7 gene in DEV-infected cells also treated with GCV and CHX, and examined DEV UL54, US2, UL13 and β-actin as controls (Gao et al., 2015; Liu et al., 2015; Hu et al., 2017). These results indicate that UL7 expression is consistent with US2 expression, which are inhibited by both GCV and CHX. Therefore, DEV UL7 gene is regulated as a γ2 gene.
Tegument proteins conserved in the herpes virus family may be involved in mediating the core process of virus assembly. HSV-1 pUL7 and pUL51 have been identified as a complex during HSV-1 replication (Roller and Fetters, 2015; Albecka et al., 2017; Feutz et al., 2019; Butt et al., 2020). pUL7 and pUL51 expression is necessary for gE localization to the junctional cell surfaces, and the complex functions in the same cell-to-cell spread pathway as gE/gI (Feutz et al., 2019). However, whether DEV pUL7 interacts with pUL51 remained unclear. Here, we showed that DEV pUL7 and pUL51 partially co-localize in the cytoplasm and nucleus of transfected cells. Moreover, Co-IP results showed that pUL7 interacted with pUL51 in co-transfected DEFs. Therefore, these results may also provide data on the pUL7/pUL51 complex to assist further research on DEV.
In conclusion, we have shown that DEV UL7 is an L gene, and its product, pUL7, is mainly localized in the cytoplasm. DEFs co-transfected with pCAGGS–HA–UL7 and pCAGGS–UL51–Flag also showed that pUL7 and pUL51 co-localized in the cytoplasm. In addition, co-IP results indicated that pUL7 interacted with pUL51 in transfected DEFs. Therefore, the interaction mechanism between pUL7 and pUL51 of DEV deserves further study.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
This study was approved by the Committee of Experimental Operational Guidelines and Animal Welfare of Sichuan Agricultural University. The experiments were conducted in accordance with approved guidelines.
Author contributions
JiH and MW designed the experiments and analyzed the data. JiH performed experiments and wrote the manuscript. AC edited the manuscript. BT provided suggestions for experiments. XO, YW, QY, DS, SZ, SM, XZ, JuH, QG, DZ, RJ, SC, and ML assisted with the experiments. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the China Agricultural Research System of MOF and MARA (CARS-42-17) and the Sichuan Veterinary Medicine and Drug Innovation Group of the China Agricultural Research System (SCCXTD-2020-18).
Acknowledgments
We thank the staff who helped with the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2024.12023/full#supplementary-material
Abbreviations
DEV, Duck enteritis virus; DP, Duck plague; PRV, Pseudorabies virus; ANT2, adenine nucleotide translocator 2; HSV-1, herpes simplex virus 1; HSV-2, herpes simplex virus 2; BHV-1, Bovine herpesvirus 1; RT-qPCR, Real-time quantitative PCR; ACV, Acyclovir; CHX, Cycloheximide; VZV, Varicella zoster virus; HCMV, Human cytomegalovirus; EBV, Epstein–Barr virus; GCV, Ganciclovir; PAA, Phosphonoacetic acid; DEFs, Duck embryo fibroblasts; MEM, Minimum essential medium; NBS, Newborn calf serum; SDS–PAGE, Sodium dodecyl sulfate–polyacrylamide gel electrophoresis; PVDF, Polyvinylidene difluoride; ECL, Chemiluminescence; WB, Western blotting; DAPI, 4′6′-diamidino-2-phenylindole; IFA, Immunofluorescence assay.
References
1
Ahlqvist J. Mocarski E. (2011). Cytomegalovirus UL103 controls virion and dense body egress. J. Virol.85 (10), 5125–5135. 10.1128/jvi.01682-10
2
Albecka A. Owen D. J. Ivanova L. Brun J. Liman R. Davies L. et al (2017). Dual function of the pUL7-pUL51 tegument protein complex in herpes simplex virus 1 infection. J. Virol.91 (2), 021966-16. 10.1128/JVI.02196-16
3
Butt B. G. Owen D. J. Jeffries C. M. Ivanova L. Hill C. H. Houghton J. W. et al (2020). Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex. Elife9 (9), e53789. 10.7554/eLife.53789
4
Deng L. Wang M. Cheng A. Yang Q. Wu Y. Jia R. et al (2020). The pivotal roles of US3 protein in cell-to-cell spread and virion nuclear egress of duck plague virus. Sci. Rep.10 (1), 7181. 10.1038/s41598-020-64190-2
5
Dhama K. Kumar N. Saminathan M. Tiwari R. Karthik K. Kumar M. A. et al (2017). Duck virus enteritis (duck plague) - a comprehensive update. Vet. Q.37 (1), 57–80. 10.1080/01652176.2017.1298885
6
Feutz E. McLeland-Wieser H. Ma J. L. Roller R. J. (2019). Functional interactions between herpes simplex virus pUL51, pUL7 and gE reveal cell-specific mechanisms for epithelial cell-to-cell spread. Virology537, 84–96. 10.1016/j.virol.2019.08.014
7
Fuchs W. Granzow H. Klopfleisch R. Klupp B. Rosenkranz D. Mettenleiter T. (2005). The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress. J. Virology79 (17), 11291–11299. 10.1128/jvi.79.17.11291-11299.2005
8
Gao J. Cheng A. C. Wang M. S. Jia R. Y. Zhu D. K. Chen S. et al (2015). Identification and characterization of the duck enteritis virus (DEV) US2 gene. Genet. Mol. Res.14 (4), 13779–13790. 10.4238/2015.October.28.40
9
Gruffat H. Marchione R. Manet E. (2016). Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front. Microbiol.7, 869. 10.3389/fmicb.2016.00869
10
He Q. Cheng A. Wang M. Xiang J. Zhu D. Zhou Y. et al (2012). Replication kinetics of duck enteritis virus UL16 gene in vitro. Virol. J.9, 281. 10.1186/1743-422x-9-281
11
He T. Wang M. Cao X. Cheng A. Wu Y. Yang Q. et al (2018). Molecular characterization of duck enteritis virus UL41 protein. Virology J.15 (1), 12. 10.1186/s12985-018-0928-4
12
He T. Wang M. Cheng A. Yang Q. Wu Y. Jia R. et al (2022). Duck plague virus UL41 protein inhibits RIG-I/MDA5-mediated duck IFN-β production via mRNA degradation activity. Veterinary Res.53 (1), 22. 10.1186/s13567-022-01043-y
13
Hu X. Wang M. Chen S. Jia R. Zhu D. Liu M. et al (2017). The duck enteritis virus early protein, UL13, found in both nucleus and cytoplasm, influences viral replication in cell culture. Poult. Sci.96 (8), 2899–2907. 10.3382/ps/pex043
14
Li H. Liu S. Han Z. Shao Y. Chen S. Kong X. (2009). Comparative analysis of the genes UL1 through UL7 of the duck enteritis virus and other herpesviruses of the subfamily Alphaherpesvirinae. Genet. Mol. Biol.32 (1), 121–128. 10.1590/s1415-47572009005000003
15
Lian B. Xu C. Cheng A. Wang M. Zhu D. Luo Q. et al (2010). Identification and characterization of duck plague virus glycoprotein C gene and gene product. Virol. J.7, 349. 10.1186/1743-422x-7-349
16
Liu C. Y. Cheng A. C. Wang M. S. Chen S. Jia R. Y. Zhu D. K. et al (2015). Duck enteritis virus UL54 is an IE protein primarily located in the nucleus. Virol. J.12, 198. 10.1186/s12985-015-0424-z
17
Liu T. Wang M. Cheng A. Jia R. Wu Y. Yang Q. et al (2020). Research Note: duck plague virus glycoprotein I influences cell-cell spread and final envelope acquisition. Poult. Sci.99 (12), 6647–6652. 10.1016/j.psj.2020.08.065
18
Masud H. Yanagi Y. Watanabe T. Sato Y. Kimura H. Murata T. (2019). Epstein-barr virus BBRF2 is required for maximum infectivity. Microorganisms7 (12), 705. 10.3390/microorganisms7120705
19
McGuire J. M. Douglas M. Smith K. D. (1996). The resolution of the neutral N-linked oligosaccharides of IgG by high pH anion-exchange chromatography. Carbohydr. Res.292, 1–9. 10.1016/s0008-6215(96)91015-0
20
Ning Y. Huang Y. Wang M. Cheng A. Jia R. Liu M. et al (2022). Evaluation of the safety and immunogenicity of duck-plague virus gE mutants. Front. Immunol.13, 882796. 10.3389/fimmu.2022.882796
21
Nozawa N. Daikoku T. Yamauchi Y. Takakuwa H. Goshima F. Yoshikawa T. et al (2002). Identification and characterization of the UL7 gene product of herpes simplex virus type 2. Virus Genes24 (3), 257–266. 10.1023/a:1015332716927
22
Plummer P. J. Alefantis T. Kaplan S. O'Connell P. Shawky S. Schat K. A. (1998). Detection of duck enteritis virus by polymerase chain reaction. Avian Dis.42 (3), 554–564. 10.2307/1592682
23
Roller R. J. Fetters R. (2015). The herpes simplex virus 1 UL51 protein interacts with the UL7 protein and plays a role in its recruitment into the virion. J. Virol.89 (6), 3112–3122. 10.1128/JVI.02799-14
24
Schmitt J. Keil G. M. (1996). Identification and characterization of the bovine herpesvirus 1 UL7 gene and gene product which are not essential for virus replication in cell culture. J. Virol.70 (2), 1091–1099. 10.1128/jvi.70.2.1091-1099.1996
25
Shen B. Li Y. Cheng A. Wang M. Wu Y. Yang Q. et al (2021). The LORF5 gene is non-essential for replication but important for duck plague virus cell-to-cell spread efficiently in host cells. Front. Microbiol.12, 744408. 10.3389/fmicb.2021.744408
26
Shen C. J. Cheng A. C. Wang M. S. Guo Y. F. Zhao L. C. Wen M. et al (2009). Identification and characterization of the duck enteritis virus UL51 gene. Arch. Virol.154 (7), 1061–1069. 10.1007/s00705-009-0407-8
27
Tanaka M. Sata T. Kawaguchi Y. (2008). The product of the Herpes simplex virus 1 UL7 gene interacts with a mitochondrial protein, adenine nucleotide translocator 2. Virol. J.5, 125. 10.1186/1743-422x-5-125
28
Wan J. Li F. Wang M. Cheng A. Tian B. Yang Q. et al (2022). The protein encoded by the duck plague virus UL14 gene regulates virion morphogenesis and affects viral replication. Poult. Sci.101 (6), 101863. 10.1016/j.psj.2022.101863
29
Wang W. Fu W. Pan D. Cai L. Ye J. Liu J. et al (2017). Varicella-zoster virus ORF7 interacts with ORF53 and plays a role in its trans-Golgi network localization. Virol. Sin.32 (5), 387–395. 10.1007/s12250-017-4048-x
30
Wu Y. Cheng A. Wang M. Zhang S. Zhu D. Jia R. et al (2011). Establishment of real-time quantitative reverse transcription polymerase chain reaction assay for transcriptional analysis of duck enteritis virus UL55 gene. Virol. J.8, 266. 10.1186/1743-422x-8-266
31
Wu Y. Cheng A. C. Wang M. S. Yang Q. Zhu D. K. Jia R. Y. et al (2012). Complete genomic sequence of Chinese virulent duck enteritis virus. J. Virol.86 (10), 5965. 10.1128/Jvi.00529-12
32
Xingli Xu S. F. Zhou J. Zhang Y. Che Y. Cai H. Wang L. et al (2016). The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of α-4 gene transcription. Virol. J.13, 152. 10.1186/s12985-016-0600-9
33
Yang L. Wang M. Cheng A. Yang Q. Wu Y. Huang J. et al (2021). UL11 protein is a key participant of the duck plague virus in its life cycle. Front. Microbiol.12, 792361. 10.3389/fmicb.2021.792361
34
Yang L. Wang M. Zeng C. Shi Y. Cheng A. Liu M. et al (2020). Duck enteritis virus UL21 is a late gene encoding a protein that interacts with pUL16. BMC Vet. Res.16 (1), 8. 10.1186/s12917-019-2228-7
35
Zhang D. Lai M. Cheng A. Wang M. Wu Y. Yang Q. et al (2017). Molecular characterization of the duck enteritis virus US10 protein. Virol. J.14 (1), 183. 10.1186/s12985-017-0841-2
36
Zhou T. Fan D. Wang M. Cheng A. Wu Y. Yang Q. et al (2021). Duck plague virus pUL48 protein activates the immediate-early gene to initiate the transcription of the virus gene. Front. Microbiol.12, 795730. 10.3389/fmicb.2021.795730
Summary
Keywords
duck enteritis virus, UL7 protein, UL51 protein, late gene, co-localization
Citation
Huang J, Wang M, Cheng A, Tian B, Ou X, Wu Y, Yang Q, Sun D, Zhang S, Mao S, Zhao X, Huang J, Gao Q, Zhu D, Jia R, Chen S and Liu M (2024) Duck enteritis virus UL7 is a late gene and the UL7-encoded protein co-localizes with pUL51. Acta Virol. 68:12023. doi: 10.3389/av.2024.12023
Received
08 September 2023
Accepted
27 June 2024
Published
16 August 2024
Volume
68 - 2024
Edited by
Ivana Nemčovičová, Slovak Academy of Sciences, Slovakia
Reviewed by
Cun Zhang, Zhejiang Academy of Agricultural Sciences, China
Igor Jurak, University of Rijeka, Croatia
Updates
Copyright
© 2024 Huang, Wang, Cheng, Tian, Ou, Wu, Yang, Sun, Zhang, Mao, Zhao, Huang, Gao, Zhu, Jia, Chen and Liu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, chenganchun@vip.163.com
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.