Abstract
Some plant RNA viruses in the family Betaflexiviridae encode a nucleic acid-binding protein (NABP) that facilitates infection by suppressing the host RNA silencing response. Previously, no members of the genus Tepovirus within this family were known to possess a NABP homolog. In this study, we identified the genome sequences of 21 novel Betaflexiviridae viruses: 17 represent new members of Tepovirus, and four may be founding members of a new genus closely related to the genus Vitivirus. Notably, five of these newly identified tepoviruses contain a NABP-like open reading frame (ORF). Sequence comparison and phylogenetic analysis of NABP homologs suggest these tepoviruses independently acquired a NABP-like ORF from diverse sources. The identification of 17 novel viruses substantially enhances our understanding of the genetic diversity within the genus Tepovirus. This study further highlights the role of recombination in the genome evolution and diversity of Betaflexiviridae.
Introduction
The Betaflexiviridae family, belonging to the order Tymovirales, comprises plant-infecting RNA viruses with monopartite positive-sense single-stranded RNA genomes, classified into two subfamilies: Trivirinae and Quinvirinae, based on their genome structures (Yoshikawa and Yaegashi, 2021). The Trivirinae subfamily comprises ten genera: Capillovirus, Chordovirus, Citrivirus, Divavirus, Prunevirus, Ravavirus, Tepovirus, Trichovirus, Vitivirus, and Wamavirus. Members of these genera typically possess three common open reading frames (ORFs): replicase (Rep), movement protein (MP), and coat protein (CP) (Vives et al., 2001; Goh et al., 2018; Goh et al., 2019). The Rep protein contains an RNA-dependent RNA polymerase (RdRp) domain responsible for viral genomic RNA replication and subgenomic RNA transcription, while MP and CP are involved in cell-to-cell movement and the encapsidation of viral genomic RNAs, respectively (Yoshikawa and Yaegashi, 2021).
The Quinvirinae subfamily contains five genera: Banmivirus, Carlavirus, Foveavirus, Robigovirus, and Sustrivirus. Members of these genera have five common ORFs: Rep, triple gene block 1 (TGB1), triple gene block 2 (TGB2), triple gene block 3 (TGB3), and CP (Park et al., 2019; Yoshikawa and Yaegashi, 2021). The three TGB proteins are required for cell-to-cell movement of the virus (Carvalho et al., 2022; Jiang et al., 2022).
Interestingly, members of the genera Carlavirus (subfamily Quinvirinae), Prunevirus, and Vitivirus (subfamily Trivirinae) often possess an additional ORF near the 3′-proximal region of their genomes (Minafra et al., 1994; Elbeaino et al., 2014; Jordan et al., 2021; Yoshikawa and Yaegashi, 2021). This terminal ORF encodes a nucleic acid-binding protein (NABP), also referred to as a cysteine-rich protein (CRP) or RNA-binding protein (RBP). These proteins have been shown to suppress the host RNA silencing response and promote viral infection (Lukhovitskaya et al., 2005; Lukhovitskaya et al., 2009; Senshu et al., 2011). NABP proteins in different genera do not exhibit significant sequence similarities, suggesting independent acquisition across lineages from unrelated sources (Goh and Hahn, 2019; Bejerman and Debat, 2022).
Recent studies have shown that some genera beyond Carlavirus, Prunevirus, and Vitivirus also harbor NABP-like ORFs. For instance, cherry mottle leaf virus (CMLV) and peach virus M (PeVM) from the genus Trichovirus, Salvia divinorum RNA virus 1 (SdRV1) from the genus Citrivirus, and Gymnadenia rhellicani virus 1 (GymRhV1) and Melampyrum roseum virus 2 (MelRoV2) from the genus Divavirus contain NABP ORFs (James et al., 2000; De La Torre-Almaraz et al., 2019; Goh and Hahn, 2019; Bejerman and Debat, 2022). These findings suggest that the acquisition of NABP—and potentially its loss—occurs relatively frequently within Betaflexiviridae (Liu et al., 2019).
Until now, no members of the genus Tepovirus were known to possess a NABP. Here, we report the first identification of novel members of the genus Tepovirus that contain a NABP homolog. These viruses were identified by systematically analyzing transcriptome data from various plants, which may have been latently infected by viruses without displaying visible symptoms. Numerous previously unknown viruses have been discovered through the analysis of assembled transcriptome contigs (Bejerman and Debat, 2022; Choi et al., 2022; Rosario et al., 2022; Shin et al., 2022a; Shin et al., 2022b; Choi and Hahn, 2023; Choi et al., 2023a; Choi et al., 2023b; Bejerman and Debat, 2024; Reddy and Sidharthan, 2024).
Materials and methods
Plant transcriptome data
To identify a virus latently infecting a hemp (Cannabis sativa) plant, we analyzed transcriptome data originally collected to study the major molecular processes underlying secondary growth and bast fiber (Behr et al., 2016; Guerriero et al., 2017; Behr et al., 2019). The plants were grown in laboratory-controlled conditions and showed no visible viral disease symptoms. The transcriptome data are available in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA435671.
To construct the extended genome sequence of Melampyrum roseum virus 2 (MelRoV2), we analyzed transcriptome data from Melampyrum roseum (SRA accession numbers DRR082664 and DRR082665) (Kado and Innan, 2018; Bejerman and Debat, 2022).
To collect plant transcriptome data potentially harboring novel tepovirus-like virus genome sequences, we utilized the Serratus Explorer1 (Edgar et al., 2022). We selected five known tepoviruses, including potato virus T (PVT), prunus virus T (PrVT), Zostera virus T (ZoVT), Trichosanthes virus A (TrVA), and Ficus tepovirus A (FiVT), as target GenBank records. We chose matches with alignment identities ranging from 60% to 90% and scores between 50 and 100. The resulting plant transcriptome datasets were further filtered to include data with an average sequence length of 100 nt or longer, paired-end layout, and Illumina platform. This process resulted in 192 plant transcriptome datasets being selected.
Viral genome identification and annotation
Raw plant transcriptome sequences were subjected to quality trimming using Sickle (version 1.33)2 with parameters “-q 30 -l 55.” The high-quality reads were then assembled into contigs using SPAdes (version 3.15.5)3 with the “rnaviral” mode (Prjibelski et al., 2020). BLASTX was used to compare contigs with known viral proteins.
Open reading frames (ORFs) in a putative virus genome contig were predicted using ORFfinder.4 Genome contigs containing complete or nearly complete Rep, MP, and CP ORFs, and showing a minimum amino acid identity of 40% with a known tepovirus Rep protein, were retained. When two or more contigs shared 90% or greater amino acid identity in the Rep proteins, only one contig was retained, and the others were discarded.
Phylogenetic analysis
Homologous viral proteins were retrieved from the NCBI protein database via the BLAST web server. We used the MAFFT online service5 to generate multiple sequence alignments under default conditions (Katoh et al., 2019). Neighbor-joining phylogenetic tree construction and bootstrap value calculation were also performed using the MAFFT online server. The resulting phylogenetic tree was visualized using MEGA (version 11.0.13)6 (Kumar et al., 2018). Visualization of multiple sequence alignments was prepared using ESPript (version 3.0)7 (Robert and Gouet, 2014).
Results
Identification of hemp virus T (HemVT) genome
Transcriptome data generated from hypocotyl tissues of young hemp plants and bast fibers isolated from adult plants were assembled into contigs (Behr et al., 2016; Guerriero et al., 2017; Behr et al., 2019). Sequence comparisons of the transcript contigs with RdRp sequences from known RNA viruses revealed contigs potentially originating from viral genomes (see Supplementary Data S1 for more detailed information). We identified a contig containing ORFs that showed significant sequence similarity to those of known members of the genus Tepovirus (subfamily Trivirinae, family Betaflexiviridae). We tentatively named this virus hemp virus T (HemVT). Its genome sequence has been deposited in the NCBI GenBank under accession number OR346818.
The HemVT genome contains four ORFs, three of which encode Rep, MP, and CP, as is typical of members of the genus Tepovirus. Notably, the fourth ORF did not exhibit amino acid sequence similarity to any known tepovirus ORFs. Instead, it showed approximately 48% sequence identity to NABP-like proteins of GymRhV1 and MelRoV2, which are members of the genus Divavirus (family Betaflexiviridae). The HemVT NABP also displayed marginal sequence identities (15%–20%) to NABP proteins of other Betaflexiviridae viruses, including those from the genera Capillovirus, Carlavirus, Citrivirus, Prunevirus, Trichovirus, and Vitivirus. Therefore, we concluded that the fourth ORF encodes a NABP-like protein, making HemVT the first member of the genus Tepovirus with a NABP homolog.
Discovery of additional tepovirus-like genomes from plant transcriptome data
Following the discovery of the HemVT genome sequence, we hypothesized that additional tepovirus genome sequences with a NABP homolog might exist in plant transcriptome data available in the NCBI SRA. To identify potential tepovirus-like genome sequences, we filtered the SRA datasets using the Serratus Explorer (Edgar et al., 2022). A total of 192 SRA datasets containing reads matching known tepovirus RdRp sequences were downloaded and assembled into contigs. We then selected contigs that contained complete or nearly complete ORFs with significant sequence identities to previously known tepovirus proteins. As a result, we identified 20 additional distinct tepovirus-like genome sequences (see Supplementary Data S2 for more detailed information). These were named according to their host plants: Acanthus hungaricus virus 1 (AcaHuV1), Allium listera virus 1 (AllLiV1), Balanophora indica virus 1 (BalInV1), Capparis spinosa virus 1 (CapSpV1), Cistanche deserticola virus 1 (CisDeV1), Crocus sativus virus 1 (CroSaV1), Davidia involucrata virus 3 (DavInV3), Ferula gummosa virus 1 (FerGuV1), Hylocereus undatus virus 1 (HylUnV1), Lilium lancifolium virus 1 (LilLaV1), Lilium pumilum virus 1 (LilPuV1), Maihueniopsis conoidea virus 1 (MaiCoV1), Panicum virgatum virus 1 (PanViV1), Pogostemon cablin virus 1 (PogCaV1), Pogostemon cablin virus 2 (PogCaV2), Solanum melongena virus 1 (SolMeV1), Vallisneria spiralis virus 1 (ValSpV1), Vallisneria spiralis virus 2 (ValSpV2), Vallisneria spiralis virus 3 (ValSpV3), and Vanilla shenzhenica virus 1 (VanShV1). The genome sequences have been deposited in the NCBI GenBank under accession numbers BK068543–BK068562 (see Supplementary Data S3, S4 for genome and protein sequences, respectively). A summary of all viruses newly identified in this study is presented in Table 1, and their genomic structures are depicted in Figure 1.
TABLE 1
| Virus | Acronym | Genus | Size (nt) | Accession | Rep (aa) | MP (aa) | CP (aa) | NABP (aa) | SRA | Host |
|---|---|---|---|---|---|---|---|---|---|---|
| Hemp virus T | HemVT | Tepovirus | 7,255 | OR346818 | 1,776 | 328 | 221 | 125 | SRR5209961 | Cannabis sativa |
| Cistanche deserticola virus 1 | CisDeV1 | Tepovirus | 7,573 | BK068547 | 1,835 | 389 | 223 | 137 | SRR10829335 | Cistanche deserticola |
| Ferula gummosa virus 1 | FerGuV1 | Tepovirus | 7,251 | BK068550 | 1,745 | 376 | 219 | 132 | SRR4428733 | Ferula gummosa |
| Maihueniopsis conoidea virus 1 | MaiCoV1 | Tepovirus | 7,295 | BK068554 | 1,739 | 356 | 204 | 139 | SRR7905848 | Maihueniopsis conoidea |
| Solanum melongena virus 1 | SolMeV1 | Tepovirus | 6,790 | BK068558 | 1,623 | 332 | 225 | 132 | SRR8736631 | Solanum melongena |
| Acanthus hungaricus virus 1 | AcaHuV1 | Tepovirus | 6,730 | BK068543 | 1,796 | 382 | >196 | SRR12034766 | Acanthus hungaricus | |
| Allium listera virus 1 | AllLiV1 | Tepovirus | 7,052 | BK068544 | 1,841 | 393 | 223 | SRR11818591 | Allium listera | |
| Balanophora indica virus 1 | BalInV1 | Tepovirus | 6,950 | BK068545 | 1,797 | 388 | 218 | SRR12009646 | Balanophora indica | |
| Capparis spinosa virus 1 | CapSpV1 | Tepovirus | 6,123 | BK068546 | 1,531 | 333 | 223 | SRR16883143 | Capparis spinosa | |
| Crocus sativus virus 1 | CroSaV1 | Tepovirus | 6,826 | BK068548 | 1,778 | 383 | 221 | SRR1140761 | Crocus sativus | |
| Davidia involucrata virus 3 | DavInV3 | Tepovirus | 6,937 | BK068549 | 1,824 | 382 | 221 | SRR2048533 | Davidia involucrata | |
| Hylocereus undatus virus 1 | HylUnV1 | Tepovirus | 6,831 | BK068551 | 1,790 | 382 | 220 | SRR7997107 | Hylocereus undatus | |
| Lilium lancifolium virus 1 | LilLaV1 | Tepovirus | 6,662 | BK068552 | 1,723 | 339 | 214 | SRR11397710 | Lilium lancifolium | |
| Lilium pumilum virus 1 | LilPuV1 | Tepovirus | 6,649 | BK068553 | 1,719 | 339 | 214 | SRR11397712 | Lilium pumilum | |
| Pogostemon cablin virus 1 | PogCaV1 | Tepovirus | 6,854 | BK068556 | 1,764 | 371 | 228 | SRR7268116 | Pogostemon cablin | |
| Pogostemon cablin virus 2 | PogCaV2 | Tepovirus | 6,803 | BK068557 | 1,792 | 382 | 221 | SRR7268115 | Pogostemon cablin | |
| Vanilla shenzhenica virus 1 | VanShV1 | Tepovirus | 6,915 | BK068562 | 1,787 | 402 | 221 | SRR5722164 | Vanilla shenzhenica | |
| Panicum virgatum virus 1 | PanViV1 | novel? | 6,623 | BK068555 | 1,705 | 336 | 202 | SRR16093774 | Panicum virgatum | |
| Vallisneria spiralis virus 1 | ValSpV1 | novel? | 6,943 | BK068559 | 1,827 | 342 | 209 | SRR16293894 | Vallisneria spiralis | |
| Vallisneria spiralis virus 2 | ValSpV2 | novel? | 6,640 | BK068560 | >1,727 | 342 | 209 | SRR16293893 | Vallisneria spiralis | |
| Vallisneria spiralis virus 3 | ValSpV3 | novel? | 7,090 | BK068561 | 1,825 | 342 | 209 | SRR16293894 | Vallisneria spiralis |
Summary of novel viruses identified in this study.
FIGURE 1
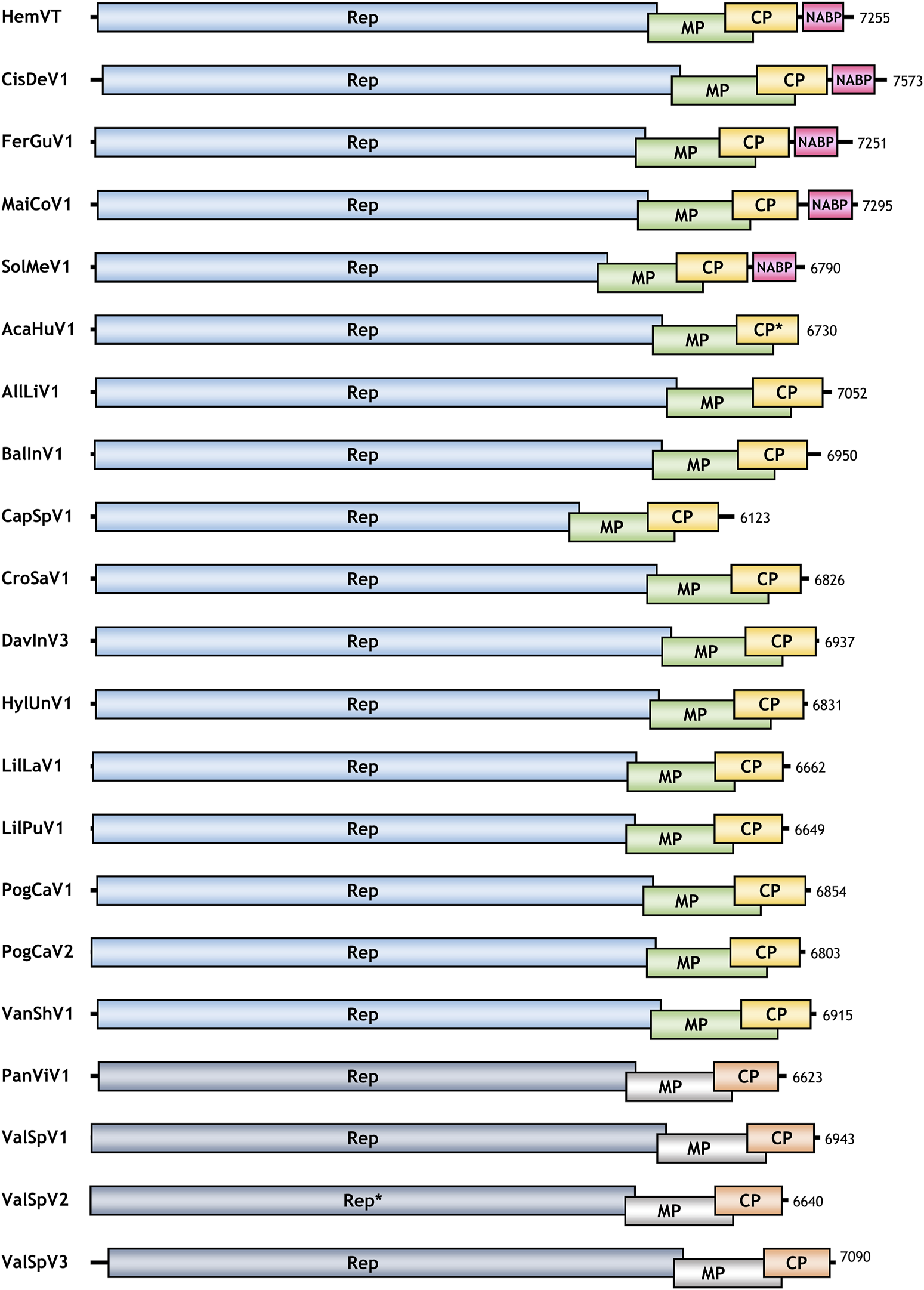
Genomic organization of viruses identified in this study. Open reading frames (ORFs) encoding replicase (Rep), movement protein (MP), coat protein (CP), and nucleic acid-binding protein (NABP) in the newly discovered viral genomes are depicted to scale. Asterisks indicate that the ORFs for the AcaHuV1 CP and ValSpV1 Rep are incomplete. ORFs of tepoviruses (top 17 viruses) and viruses that may belong to a novel genus (bottom four viruses) are shaded in different colors.
Upon examining the genome organization of the newly identified viruses, we found that, in addition to HemVT, four more viruses—CisDeV1, FerGuV1, MaiCoV1, and SolMeV1—contained a NABP-like ORF in their genomes. The other 16 viruses (AcaHuV1, AllLiV1, BalInV1, CapSpV1, CroSaV1, DavInV3, HylUnV1, LilLaV1, LilPuV1, PogCaV1, PogCaV2, VanShV1, PanViV1, ValSpV1, ValSpV2, and ValSpV3) did not contain a NABP ORF. In the case of AcaHuV1, the presence of the fourth ORF could not be determined because its genome sequence was truncated in the middle of the CP ORF.
Phylogenetic analysis of newly identified tepovirus-like genomes
To establish the phylogenetic relationships of the newly identified viruses and known Tepovirus species, a phylogenetic tree was constructed (Figure 2). We collected the Rep protein sequences of all nine known tepoviruses and representative viruses from other genera in the family Betaflexiviridae. A multiple sequence alignment was generated and a phylogenetic tree was inferred using the MAFFT online service (Katoh et al., 2019). Among the 21 newly identified viruses, 17 (AcaHuV1, AllLiV1, BalInV1, CapSpV1, CisDeV1, CroSaV1, DavInV3, FerGuV1, HemVT, HylUnV1, LilLaV1, LilPuV1, MaiCoV1, PogCaV1, PogCaV2, SolMeV1, and VanShV1) formed a clade with the known tepoviruses, suggesting that they belong to the genus Tepovirus. However, this clade was weakly supported, with a bootstrap value of 36, consistent with previous findings (Goh et al., 2021).
FIGURE 2
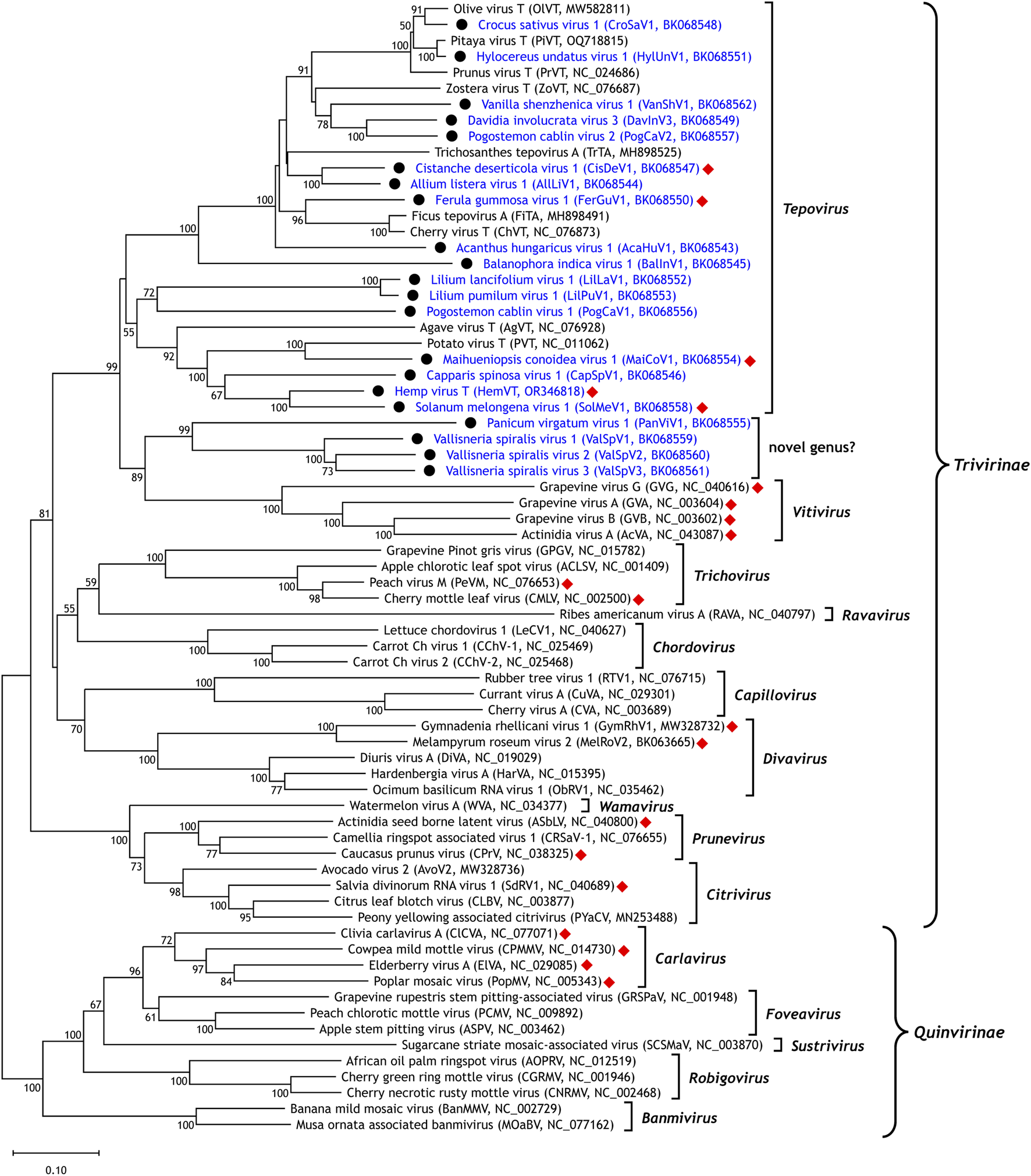
Phylogenetic relationships of viruses identified in this study. A phylogenetic tree was constructed based on a multiple alignment of replicase (Rep) protein sequences from the newly discovered viruses (marked with a black circle), nine known members of the genus Tepovirus, and representative members of other genera within the family Betaflexiviridae. Among the newly identified viruses, 17 are grouped within the genus Tepovirus, while four form a separate subclade, potentially representing a novel genus closely related to the genus Vitivirus. Bootstrap support values of 50 or greater are shown. Viruses possessing a nucleic acid-binding protein (NABP) are indicated by a red diamond.
All five viruses containing a NABP homolog (CisDeV1, FerGuV1, HemVT, MaiCoV1, and SolMeV1) were positioned within the Tepovirus clade, confirming the discovery of tepoviruses with NABP-like ORFs. However, the other 12 newly identified tepoviruses lacked the fourth ORF, indicating that most tepoviruses do not possess a NABP-like ORF.
The remaining four newly identified viruses (PanViV1, ValSpV1, ValSpV2, and ValSpV3) formed a distinct subclade with strong bootstrap support (99). This clade is closely related to the genus Vitivirus but does not contain a NABP-like ORF, whereas members of Vitivirus are known to possess NABPs. The absence of a NABP-like ORF and the formation of a distinct clade suggest that these four viruses may represent the founding members of a novel genus closely related to Vitivirus.
Phylogenetic analysis of nucleic acid-binding proteins in newly identified tepoviruses
Next, we performed sequence comparison and phylogenetic analysis of the five newly discovered tepovirus NABP homologs (Figure 3). First, we searched the NCBI protein database using the five tepovirus NABP homologs as queries. The BLASTP search used an E-value threshold of 1e−5, and 28 known NABP proteins showing sequence similarities to the tepovirus NABP-like proteins were retrieved.
FIGURE 3
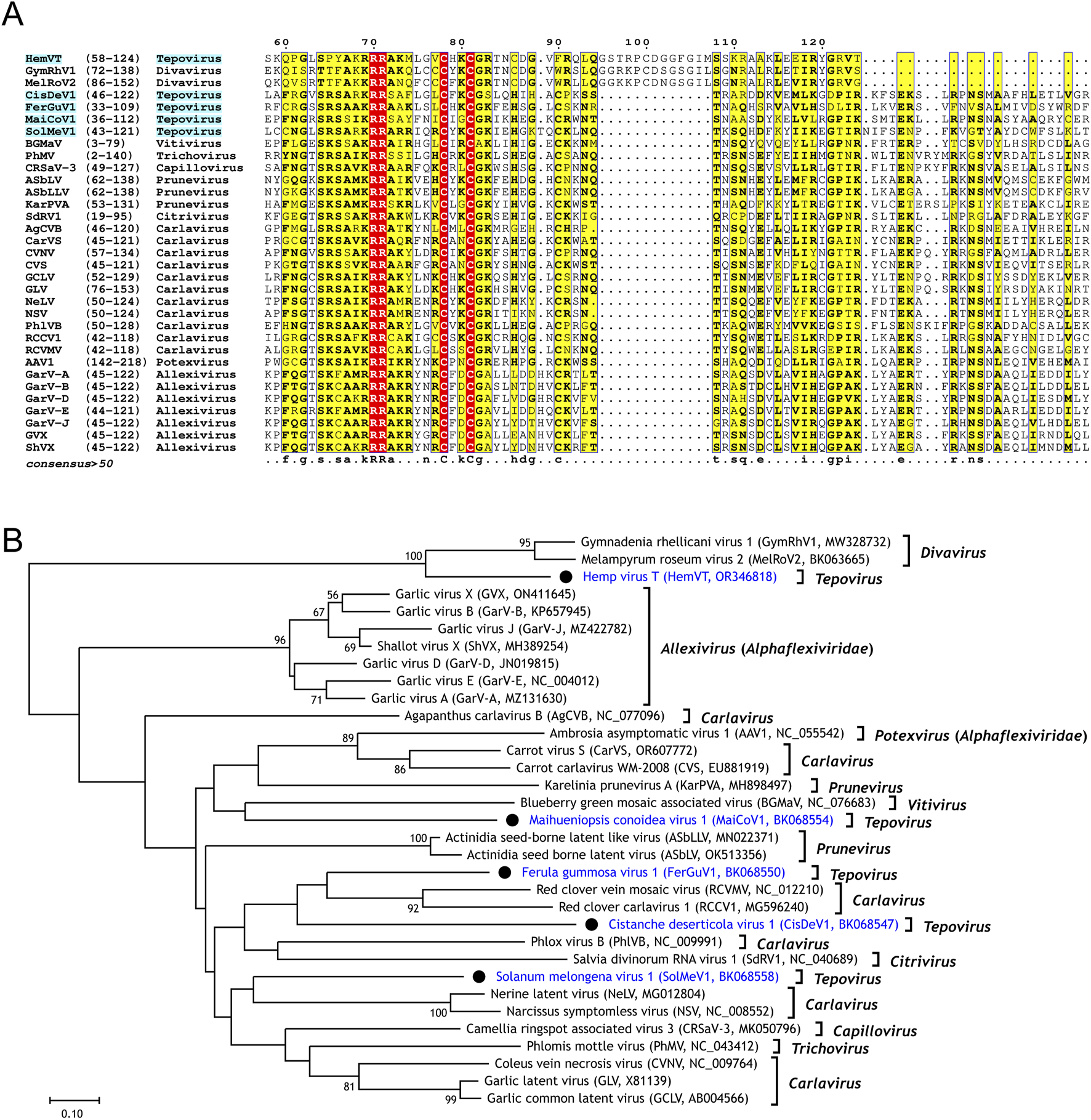
Sequence comparison and phylogenetic analysis of nucleic acid-binding proteins (NABPs). (A) An excerpt from the multiple sequence alignment of NABP homologs from five newly identified tepoviruses (highlighted in cyan) and related viruses is displayed. Identical residues across all sequences and those conserved in half or more of the sequences are highlighted with red and yellow backgrounds, respectively. Amino acid coordinates are provided in parentheses. See Supplementary Figure S1 for the full sequence alignment. (B) A phylogenetic tree constructed from a multiple alignment of NABP protein sequences from novel tepoviruses (marked with a black circle) and related viruses is presented. Bootstrap support values of 50 or greater are shown. Virus genome acronyms and NCBI accession numbers are included in parentheses.
A multiple sequence alignment of the five tepovirus NABP-like proteins and the 28 known NABPs revealed two distinct groups (Figure 3A). The first group included HemVT, GymRhV1, and MelRoV2. The HemVT NABP-like protein shared approximately 48% identity with the NABPs of GymRhV1 and MelRoV2, both members of the genus Divavirus. The second group consisted of the remaining viruses, including four tepoviruses (CisDeV1, FerGuV1, MaiCoV1, and SolMeV1), and 26 known viruses. The four tepovirus NABP-like proteins exhibited 12%–36% identity with previously known NABP proteins. Among the 26 known viruses, 18 belonged to six genera (Capillovirus, Carlavirus, Citrivirus, Prunevirus, Trichovirus, and Vitivirus) within Betaflexiviridae. Notably, eight of the 26 viruses were from two genera (Allexivirus and Potexvirus) in the family Alphaflexiviridae.
The phylogenetic tree inferred from the multiple alignment of NABP homolog sequences confirmed that the HemVT NABP homolog shares ancestry with those of GymRhV1 and MelRoV2 (Figure 3B). The strong bootstrap support (100) for this clade, along with their high sequence similarity, suggests that HemVT, GymRhV1, and MelRoV2 recently obtained their NABP-like ORFs from closely related sources. In the case of GymRhV1 and MelRoV2, it is more plausible that the NABP-like ORF was acquired in their common ancestor before their divergence, as both their Rep and NABP-like proteins show high sequence similarity.
The phylogenetic tree also showed that the NABP-like proteins of CisDeV1, FerGuV1, MaiCoV1, and SolMeV1 share ancestry with those from other genera in Betaflexiviridae and Alphaflexiviridae. However, the exact phylogenetic relationships remain unclear due to low bootstrap support values for the subclades containing them. This ambiguous relationship suggests that these viruses may have acquired their NABP-like ORFs from unrelated sources. This explanation is further supported by the discordance between the Rep and NABP phylogenetic trees. For example, in the Rep tree, MaiCoV1, SolMeV1, and HemVT form a strongly supported subclade (bootstrap value of 100), with HemVT being the closest relative to SolMeV1. However, in the NABP tree, MaiCoV1 and SolMeV1 are distantly placed, and HemVT possesses an NABP-like protein that is distinct from those found in other related viruses. Therefore, it is highly likely that these viruses independently obtained their NABP-like ORFs from unrelated sources.
Discussion
RNA viruses must evade the host RNA silencing response, which is triggered by viral double-stranded RNAs (Roth et al., 2004). Core viral proteins involved in replication, movement, and encapsulation of viral genomic RNAs are often recruited to function as suppressors of RNA silencing (Park et al., 2013; Bellott et al., 2019). In some cases, viruses encode a specific protein, such as the NABP found in certain members of the family Betaflexiviridae, which has been associated with the suppression of RNA silencing (Lukhovitskaya et al., 2005; Lukhovitskaya et al., 2009; Senshu et al., 2011).
Previously, NABP ORFs were identified in only three Betaflexiviridae genera: Carlavirus, Prunevirus, and Vitivirus (Minafra et al., 1994; Elbeaino et al., 2014; Jordan et al., 2021; Yoshikawa and Yaegashi, 2021). However, as more genomes have been identified, NABP-like ORFs have also been identified in other genera. For instance, CMLV and PeVM (Trichovirus), SdRV1 (Citrivirus), and GymRhV1 and MelRoV2 (Divavirus) contain NABP-like ORFs (James et al., 2000; De La Torre-Almaraz et al., 2019; Goh and Hahn, 2019; Bejerman and Debat, 2022). Interestingly, camellia ringspot associated virus 1 (CRSaV-1), a Prunevirus member, lacks a NABP ORF, despite NABP being considered characteristic of this genus (Liu et al., 2019). This indicates that NABP genes may act as accessory elements that may be gained or lost through recombination events.
The presence of two distinct types of NABP homologs among the newly identified tepoviruses suggests independent acquisition from unrelated sources. Prior research has demonstrated that recombination events are common in Betaflexiviridae and play a significant role in the evolution of viral genomes (Martelli et al., 2007; Alabi et al., 2014; Marais et al., 2015; Yoshikawa and Yaegashi, 2021; Silva et al., 2022). Our findings support the idea that recombination events involving NABP-like ORFs occur frequently within Betaflexiviridae. The acquisition of NABP homologs may provide viruses with an advantage in evading host defenses, particularly through the suppression of RNA silencing. NABPs may also have additional functions, as many viral proteins are known to perform multiple roles during infection (Bellott et al., 2019). Although sequence similarity and genomic organization suggest that the newly identified NABP homologs in tepoviruses could act as suppressors of RNA silencing, their precise functions require experimental validation.
In this study, we identified 21 novel RNA viruses, 17 of which are new members of the genus Tepovirus. To date, only nine tepovirus genome sequences have been reported, and five of these are officially recognized by the International Committee on Taxonomy of Viruses (ICTV)8. This work substantially increases the known diversity of the genus Tepovirus. Additionally, we identified four viruses that may represent a new genus closely related to Vitivirus. The genome sequences identified here provide valuable insights into the evolutionary processes influencing the Betaflexiviridae family.
Statements
Data availability statement
The viral genome sequences identified in this study have been deposited in NCBI GenBank under the accession numbers OR346818, BK063665, and BK068543–BK068562, and are also included in the article/Supplementary Material.
Author contributions
SL, GG, and J-FH obtained the hemp transcriptome data; DC, HP, SB, MSC, and YH performed bioinformatics analyses; YH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) through grants funded by the Government of Korea (grant numbers 2018R1A5A1025077 and RS-2023-00208564).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2024.13952/full#supplementary-material
Footnotes
2.^ https://github.com/najoshi/sickle
3.^ https://github.com/ablab/spades
4.^ https://www.ncbi.nlm.nih.gov/orffinder
References
1
Alabi O. J. Al Rwahnih M. Mekuria T. A. Naidu R. A. (2014). Genetic diversity of grapevine virus A in Washington and California vineyards. Phytopathology104, 548–560. 10.1094/PHYTO-06-13-0179-R
2
Behr M. Legay S. Zizkova E. Motyka V. Dobrev P. I. Hausman J. F. et al (2016). Studying secondary growth and bast fiber development: The hemp hypocotyl peeks behind the wall. Front. Plant Sci.7, 1733. 10.3389/fpls.2016.01733
3
Behr M. Lutts S. Hausman J. F. Sergeant K. Legay S. Guerriero G. (2019). De novo transcriptome assembly of textile hemp from datasets on hypocotyls and adult plants. Data Brief.27, 104790. 10.1016/j.dib.2019.104790
4
Bejerman N. Debat H. (2022). Exploring the tymovirales landscape through metatranscriptomics data. Arch. Virol.167, 1785–1803. 10.1007/s00705-022-05493-9
5
Bejerman N. Debat H. (2024). Alpha- and Betagymnorhavirus: two new genera of gymnosperm-infecting viruses in the family Rhabdoviridae, subfamily Betarhabdovirinae. Arch. Virol.169, 193. 10.1007/s00705-024-06119-y
6
Bellott L. Gilmer D. Michel F. (2019). Hit two birds with one stone: The multiple properties of (viral) RNA silencing suppressors. Virol. (Montrouge)23, 38–60. 10.1684/vir.2019.0798
7
Carvalho S. L. Tilsner J. Figueira A. R. Carvalho C. M. (2022). Subcellular localization and interactions among TGB proteins of cowpea mild mottle virus. Arch. Virol.167, 2555–2566. 10.1007/s00705-022-05576-7
8
Choi D. Hahn Y. (2023). Quantitative analysis of RNA polymerase slippages for production of P3N-PIPO trans-frame fusion proteins in potyvirids. J. Microbiol.61, 917–927. 10.1007/s12275-023-00083-z
9
Choi D. Rai M. Rai A. Shin C. Yamazaki M. Hahn Y. (2023a). High-throughput RNA sequencing analysis of Mallotus japonicus revealed novel polerovirus and amalgavirus. Acta Virol.67, 13–23. 10.4149/av_2023_102
10
Choi D. Rai M. Rai A. Yamazaki M. Hahn Y. (2023b). Discovery of two novel potyvirus genome sequences by high-throughput RNA sequencing in Aconitum carmichaelii tissue samples. Acta Virol.67, 11782. 10.3389/av.2023.11782
11
Choi D. Shin C. Shirasu K. Ichihashi Y. Hahn Y. (2022). Artemisia capillaris nucleorhabdovirus 1, a novel member of the genus Alphanucleorhabdovirus, identified in the Artemisia capillaris transcriptome. Acta Virol.66, 149–156. 10.4149/av_2022_204
12
De La Torre-Almaraz R. Pallas V. Sanchez-Navarro J. A. (2019). Molecular characterization of a new trichovirus from peach in Mexico. Arch. Virol.164, 2617–2620. 10.1007/s00705-019-04358-y
13
Edgar R. C. Taylor J. Lin V. Altman T. Barbera P. Meleshko D. et al (2022). Petabase-scale sequence alignment catalyses viral discovery. Nature602, 142–147. 10.1038/s41586-021-04332-2
14
Elbeaino T. Giampetruzzi A. De Stradis A. Digiaro M. (2014). Deep-sequencing analysis of an apricot tree with vein clearing symptoms reveals the presence of a novel betaflexivirus. Virus Res.181, 1–5. 10.1016/j.virusres.2013.12.030
15
Goh C. J. Hahn Y. (2019). Identification of a novel member of the family Betaflexiviridae from the hallucinogenic plant Salvia divinorum. Acta Virol.63, 373–379. 10.4149/av_2019_401
16
Goh C. J. Park D. Hahn Y. (2021). A novel tepovirus, Agave virus T, identified by the analysis of the transcriptome data of blue agave (Agave tequilana). Acta Virol.65, 68–71. 10.4149/av_2021_107
17
Goh C. J. Park D. Kim H. Sebastiani F. Hahn Y. (2018). Novel Divavirus (the family Betaflexiviridae) and Mitovirus (the family Narnaviridae) species identified in basil (Ocimum basilicum). Acta Virol.62, 304–309. 10.4149/av_2018_224
18
Goh C. J. Park D. Lee J. S. Davey P. A. Pernice M. Ralph P. J. et al (2019). Zostera virus T - a novel virus of the genus Tepovirus identified in the eelgrass, Zostera muelleri. Acta Virol.63, 366–372. 10.4149/av_2019_409
19
Guerriero G. Behr M. Legay S. Mangeot-Peter L. Zorzan S. Ghoniem M. et al (2017). Transcriptomic profiling of hemp bast fibres at different developmental stages. Sci. Rep.7, 4961. 10.1038/s41598-017-05200-8
20
James D. Jelkmann W. Upton C. (2000). Nucleotide sequence and genome organisation of cherry mottle leaf virus and its relationship to members of the Trichovirus genus. Arch. Virol.145, 995–1007. 10.1007/s007050050690
21
Jiang C. Shan S. Huang Y. Mao C. Zhang H. Li Y. et al (2022). The C-terminal transmembrane domain of cowpea mild mottle virus TGBp2 is critical for plasmodesmata localization and for its interaction with TGBp1 and TGBp3. Front. Microbiol.13, 860695. 10.3389/fmicb.2022.860695
22
Jordan R. Korolev E. Grinstead S. Mollov D. (2021). First complete genome sequence of carnation latent virus, the type member of the genus Carlavirus. Arch. Virol.166, 1501–1505. 10.1007/s00705-021-04999-y
23
Kado T. Innan H. (2018). Horizontal gene transfer in five parasite plant species in Orobanchaceae. Genome Biol. Evol.10, 3196–3210. 10.1093/gbe/evy219
24
Katoh K. Rozewicki J. Yamada K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform.20, 1160–1166. 10.1093/bib/bbx108
25
Kumar S. Stecher G. Li M. Knyaz C. Tamura K. (2018). Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35, 1547–1549. 10.1093/molbev/msy096
26
Liu H. Wu L. Zheng L. Cao M. Li R. (2019). Characterization of three new viruses of the family Betaflexiviridae associated with camellia ringspot disease. Virus Res.272, 197668. 10.1016/j.virusres.2019.197668
27
Lukhovitskaya N. I. Ignatovich I. V. Savenkov E. I. Schiemann J. Morozov S. Y. Solovyev A. G. (2009). Role of the zinc-finger and basic motifs of chrysanthemum virus B p12 protein in nucleic acid binding, protein localization and induction of a hypersensitive response upon expression from a viral vector. J. Gen. Virol.90, 723–733. 10.1099/vir.0.005025-0
28
Lukhovitskaya N. I. Solovyev A. G. Koshkina T. E. Zavriev S. K. Morozov S. (2005). Interaction of cysteine-rich protein of Carlavirus with plant defense system. Mol. Biol. Mosk.39, 896–904. 10.1007/s11008-005-0094-9
29
Marais A. Faure C. Mustafayev E. Candresse T. (2015). Characterization of new isolates of apricot vein clearing-associated virus and of a new Prunus-infecting virus: evidence for recombination as a driving force in Betaflexiviridae evolution. PLoS One10, e0129469. 10.1371/journal.pone.0129469
30
Martelli G. P. Adams M. J. Kreuze J. F. Dolja V. V. (2007). Family Flexiviridae: A case study in virion and genome plasticity. Annu. Rev. Phytopathol.45, 73–100. 10.1146/annurev.phyto.45.062806.094401
31
Minafra A. Saldarelli P. Grieco F. Martelli G. P. (1994). Nucleotide sequence of the 3' terminal region of the RNA of two filamentous grapevine viruses. Arch. Virol.137, 249–261. 10.1007/BF01309473
32
Park D. Zhang M. Hahn Y. (2019). Novel Foveavirus (the family Betaflexiviridae) species identified in ginseng (Panax ginseng). Acta Virol.63, 155–161. 10.4149/av_2019_204
33
Park M. R. Seo J. K. Kim K. H. (2013). Viral and nonviral elements in potexvirus replication and movement and in antiviral responses. Adv. Virus Res.87, 75–112. 10.1016/B978-0-12-407698-3.00003-X
34
Prjibelski A. Antipov D. Meleshko D. Lapidus A. Korobeynikov A. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinforma.70, e102. 10.1002/cpbi.102
35
Reddy T. S. Sidharthan V. K. (2024). Three-fold expansion of the genetic diversity of blunerviruses through plant (meta)transcriptome data-mining. Virology599, 110210. 10.1016/j.virol.2024.110210
36
Robert X. Gouet P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res.42, W320–W324. 10.1093/nar/gku316
37
Rosario K. Van Bogaert N. Lopez-Figueroa N. B. Paliogiannis H. Kerr M. Breitbart M. (2022). Freshwater macrophytes harbor viruses representing all five major phyla of the RNA viral kingdom Orthornavirae. PeerJ10, e13875. 10.7717/peerj.13875
38
Roth B. M. Pruss G. J. Vance V. B. (2004). Plant viral suppressors of RNA silencing. Virus Res.102, 97–108. 10.1016/j.virusres.2004.01.020
39
Senshu H. Yamaji Y. Minato N. Shiraishi T. Maejima K. Hashimoto M. et al (2011). A dual strategy for the suppression of host antiviral silencing: Two distinct suppressors for viral replication and viral movement encoded by potato virus M. J. Virol.85, 10269–10278. 10.1128/JVI.05273-11
40
Shin C. Choi D. Shirasu K. Hahn Y. (2022a). Identification of dicistro-like viruses in the transcriptome data of Striga asiatica and other plants. Acta Virol.66, 157–165. 10.4149/av_2022_205
41
Shin C. Choi D. Shirasu K. Ichihashi Y. Hahn Y. (2022b). A novel RNA virus, Thesium chinense closterovirus 1, identified by high-throughput RNA-sequencing of the parasitic plant Thesium chinense. Acta Virol.66, 206–215. 10.4149/av_2022_302
42
Silva J. M. F. Melo F. L. Elena S. F. Candresse T. Sabanadzovic S. Tzanetakis I. E. et al (2022). Virus classification based on in-depth sequence analyses and development of demarcation criteria using the Betaflexiviridae as a case study. J. Gen. Virol.103, 001806. 10.1099/jgv.0.001806
43
Vives M. C. Galipienso L. Navarro L. Moreno P. Guerri J. (2001). The nucleotide sequence and genomic organization of citrus leaf blotch virus: Candidate type species for a new virus genus. Virology287, 225–233. 10.1006/viro.2001.1040
44
Yoshikawa N. Yaegashi H. (2021). “Betaflexiviruses (Betaflexiviridae),” in Encyclopedia of virology. Editors BamfordD. H.ZuckermanM.4th ed. (Oxford: Academic Press), 229–238. 10.1016/B978-0-12-809633-8.21332-0
Summary
Keywords
nucleic acid-binding protein (NABP), Tepovirus , Betaflexiviridae , virus genome evolution, plant virus
Citation
Choi D, Park H, Baek S, Choi MS, Legay S, Guerriero G, Hausman J-F and Hahn Y (2025) Discovery of novel tepovirus genomes with a nucleic acid-binding protein homolog by systematic analysis of plant transcriptome data. Acta Virol. 68:13952. doi: 10.3389/av.2024.13952
Received
17 October 2024
Accepted
30 December 2024
Published
10 January 2025
Volume
68 - 2024
Edited by
Zdeno Šubr, Slovak Academy of Sciences, Slovakia
Reviewed by
Adrian Alejandro Valli, Spanish National Research Council (CSIC), Spain
Lukas Predajna, Slovak Academy of Sciences, Slovakia
Deya Wang, Zaozhuang University, China
Updates
Copyright
© 2025 Choi, Park, Baek, Choi, Legay, Guerriero, Hausman and Hahn.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoonsoo Hahn, hahny@cau.ac.kr
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.